
Familial
and late-onset Alzheimer’s disease: Evidence for an auto-immune component
triggered by viral, microbial and allergen antigens with homology to beta-amyloid
and APP mutant peptides. by Chris
Carter is licensed under a Creative
Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License.
C.J.Carter:
Also published in pdf format at NaturePrecedings
Enhanced links powered by

Flat 4, 20 Upper Maze Hill, St Leonard’s on Sea East Sussex
Abstract
Autoantibodies to beta-amyloid are common in the ageing population
and in Alzheimer’s disease and may derive from aberrant forms of beta-amyloid
(Abeta) or from antigens with homology to it. The latter seems likely, as
glycoprotein B of the Herpes simplex virus (HSV-1) and 69 other viruses and
phages, including HHV-6, Hepatitis C, polyoma viruses and HIV-1 show exact
homology with a VGGVV C-terminal fibrillogenic sequence, an epitope that labels
beta-amyloid in the Alzheimer’s disease brain. Other viruses, including those
causing the common cold and influenza also show significant homology with
known and predicted beta amyloid epitopes, as do a number of allergens (food,
pollen and insect venoms and most notably house dust mites). Bacteria implicated
as risk factors in Alzheimer’s disease (C.Pneumoniae, B.Burgdorferri , H.Pylori,
P.Gingivalis) also contain matching Abeta proteins as does the meningitis
causing fungus, C.Neoformans, which has been associated with a rare but curable
misdiagnosed form of Alzheimer’s disease. Immune activation occurs in the
Alzheimer’s disease brain, as evidenced by many immune-related proteins in
plaques and by the presence of the complement membrane attack complex in neurones.
These observations suggest that Alzheimer’s disease is an autoimmune disorder
triggered by pathogens with homology to beta-amyloid, whose antibodies target
and kill the neurones within which the peptide resides, via immune and inflammatory
activation and complement-related lysis. Viruses matching the Abeta target
regions of beneficial catalytic autoantibodies include those from components
of the Mediterranean diet (plant viruses) and from the papillomavirus and
other cancer-inducing viruses, both of which (cancer and diet) are inversely
associated with Alzheimer’s disease risk. As a vaccine against the human papillomavirus
already exists, it may have a role to play in the prevention of Alzheimer’s
disease. This scenario explains most of the epidemiological observations in
Alzheimer’s disease which is more common in women and Afro-Americans, as is
HSV-2 seroprevalence; related to the number of pregnancies and thus greater
exposure to childhood infections, and less severe in nuns, who are shielded
from sexually transmitted diseases. Atopy and autoimmune disorders are common
in Alzheimer’s disease in accord with allergen homology to Abeta, and the
use of anti-inflammatory agents has been reported to reduce the risk of developing
Alzheimer’s disease. HIV-1 infection can cause dementia with Alzheimer’s disease
pathology, again in accord with Abeta homology. The four major risk genes
in Alzheimer’s disease, APOE, clusterin, complement receptor 1 and PICALM
can all be related to viral life cycles and clusterin and complement receptor
1 are both complement inhibitors. This scenario is also relevant to familial
Alzheimer’s disease, as the four mutant forms of APP717 and the
Swedish mutant (APP670/671) convert the surrounding peptide to
matches with commensal bacterial flora (E.Coli, E. Faecalis, P.Gingivalis)
and to viruses with high seroprevalence (HHV-6, norovirus and polyoma viruses)
and to those causing influenza and the common cold. Multiple lines of evidence
thus suggest that late-onset and Familial Alzheimer’s disease are autoimmune
disorders caused by diverse common pathogens and allergens that are homologous
to beta-amyloid or mutant APP fragments. Antibodies to these multiple agents
are a likely source of the autoantibodies to beta-amyloid in the ageing population
and would accumulate with repeated antigen exposure and age, the greatest
risk factor in Alzheimer’s disease. Vaccination, which has already been shown
to reduce the incidence of Alzheimer’s disease (diphtheria, influenza, tetanus,
and polio), pathogen screening and elimination, and immunosuppressant therapy
may be of therapeutic benefit in this disorder. This scenario is also relevant
to several other autoimmune disorders including multiple sclerosis, myasthenia
gravis, pemphigus vulgaris, systemic lupus erythematosus, and Chronic obstructive
pulmonary disease where the known autoantigens also line up with the reported
viral risk factors. Mutant proteins from other genetic diseases (Huntington’s
disease and other PolyGlutamine repeat disorders) and cystic fibrosis also
align with very common viruses. This may well be a near universal phenomenon,
reflecting the idea that all life evolved from viruses, which have however
left behind a deadly legacy of human viral derived proteins with homology
to antigenic regions of the current virome. This could well be responsible
for most of our ills. This also suggests that vaccination using epitopes to
the non-homologous regions of the viral culprits may be of benefit in autoimmune
and even in human genetic disorders.
Introduction
Autoantibodies to beta-amyloid are common in the ageing population
and in Alzheimer’s disease and may exert a beneficial role in adsorbing the
toxic peptide or catalysing its destruction. They may also mount an immune
attack against beta-amyloid, activating inflammatory pathways and complement
cascades that kill the neurones in which the peptide resides (1-3) . The membrane
attack complex of the complement pathway is present in Alzheimer’s disease
neurones (4;5) , supporting a role for aberrant immune/complement activation
within the brain. The source of these autoantibodies is not clear. They could
be derived as a response to abnormal forms of beta-amyloid or from antibodies
to other antigens that cross-react with the peptide.
It has already been noted that glycoprotein B of the Herpes simplex
virus shows marked homology with beta-amyloid, particularly matching a VGGVV
C-terminus pentapeptide (6) (Fig 1). The VGGVG epitope has been used to label
beta-amyloid1-40 in extracellular neurofibrillary tangles (7) .
This pentapeptide, per se, forms aggregates characterised by twisted
ropes and banded fibrils (8) . This is a characteristic of both beta-amyloid
and of HSV-1 glycoprotein B peptide fragments containing this sequence. The
viral glycoprotein B fragments form thioflavine T positive fibrils which accelerate
beta-amyloid fibril formation, and are neurotoxic in cell culture (9) .Herpes
simplex infection (HSV-1) has been shown to be a risk factor in Alzheimer’s
disease, acting in synergy with possession of the APOE4 allele (10) . HSV-1
infection in mice or neuroblastoma cells increases beta-amyloid deposition
and phosphorylation of the microtubule protein tau (11;12) .Viral infection
in mice also results in hippocampal and entorhinal cortex neuronal degeneration
and memory loss, all as found in Alzheimer’s disease (13) . A recent study
has also shown that anti-HSV-1 immunoglobulin M seropositivity, a marker of
primary viral infection or reactivation, in a cohort of healthy patients,
was significantly associated with the subsequent development of Alzheimer’s
disease. Anti-HSV-1 IgG, a marker of lifelong infection showed no association
with subsequent Alzheimer’s disease development (14) . All of these factors
support a viral influence on the development of Alzheimer’s disease. Antibodies
to the Potato virus Y, which is highly homologous to beta-amyloid (Fig 1),
are also able to label beta-amyloid containing plaques in the Alzheimer’s
disease brain (15) . It is therefore possible that beta-amyloid autoantibodies
are derived from such homologous antigens. Homology searches within viral
and microbial proteomes and allergenic proteins showed that many are highly
homologous to immunogenic regions of the beta-amyloid peptide and also that
APP mutations in Familial Alzheimer’s disease convert the surrounding peptides
to matches to very common viruses and bacteria.
Methods
Homology searches against viral, bacterial or fungal proteomes
were performed via the NCBI or Uniprot BLAST servers and sequence alignments
via the CLUSTAL server at UniProt (16) . Homology with allergen sequences
was determined by interrogation of the Structural database of Allergenic Proteins
(17) http://fermi.utmb.edu/SDAP/index.html
and from homology searches at the AllergenOnLine database http://www.allergenonline.org/ (18)
. Epitopes containing viral/beta amyloid matching sequences were found using
the Human Immune epitope database. www.immuneepitope.org
(19) . B-Cell epitopes were identified using the BepiPred server http://www.cbs.dtu.dk/services/BepiPred/
(20) .
Results
The homology between HSV-1 glycoprotein B and beta-amyloid1-42
is shown in Fig 1. A homology search against viral proteomes showed
that a number of other viruses contain this VGGVV sequence (Table 1). These
include adenovirus 8, Dengue virus, Herpes Simplex (HSV-1, 2 and 6) hepatitis
C, Lactate dehydrogenase-elevating virus, the polyoma virus and HIV-1, viruses
infecting family pets (cats, guinea pigs and goldfish) and farmyard animals
(cattle, horses, pigs and poultry) and viruses infecting certain foodstuffs
(cherries, radish, strawberries and raspberries, tomatoes, potatoes, watermelon,
oysters, salmon and shrimp). The VVGGV sequence is also present in a number
of phages infecting bacteria that cause common childhood and adult diseases
(Gastroenteritis, food poisoning, hospital infections (nococosmial), and wound
infections). It should be noted that the VGGVV sequence was restricted to
69 viruses and phages (Table 1) out of 2463 viral genomes in the NCBI database.
A further homology search against the viral proteome
database revealed that many other common viruses express proteins with marked
homology (pentapeptides or more) to other sequences within the beta-amyloid
peptide (Table 2). These viruses include Adenovirus D, Coxsackie virus B2,
influenza, hepatitis C and Yellow fever, numerous HIV-1 or HIV-2 proteins,
and the human papillomavirus. Bovine and poultry viruses were also well represented.
Phages infecting campylobacter, enterobacteria, lactococcus, mycobacterium,
streptococci and vibrio (Pentapeptide or more) as well as Listeria and pseudomonas
(Tetrapeptide matches) were also represented . These infect very common benign
or infection-related bacteria. Although less homologous, viruses causing the
common cold, mumps, rubella and polio nevertheless contained exactly matching
tetrapeptide beta-amyloid sequences.
Numerous plant viruses contain both the VGGVV
and other matching beta-amyloid sequences. Plant viruses are generally considered
as benign or unable to infect humans. However, they are obviously ingested
and exist in human faeces (21) . The Pepper mottle virus has recently been
associated with fever in Man (22) suggesting that phytonosis might be more
common than thought. A capsid protein of this virus contains the YEVHHQ sequence
of beta-amyloid as do a number of other plant viruses, including the potato
virus. This sequence is immunogenic (in beta-amyloid) and antibodies to the
potato virus label amyloid plaques in the Alzheimer’s disease brain (15) .
Sequences within the potato virus polyprotein show 54% identity with beta-amyloid1-28
(Fig 1).
Many of the viruses containing beta-amyloid sequences
are very common (eg influenza and Herpes viruses and even the common cold
virus) and the beta-amyloid matching phages are associated with a number of
bacteria causing common gastrointestinal problems (e.g. clostridum, salmonella
and vibrio) or associated with hospital infections (Pseudomonas, Serratia).
Exposure to these agents, which possess myriad strains, is ineluctable and
each exposure is likely to increase the risk of generating antibodies that
might also target the beta-amyloid peptide.
Bacteria and Fungi implicated in Alzheimer’s disease
The VGGVV sequence, and others, are also present in proteins
from C.Pneumoniae, Borrelia Burgdorferri and Helicobacter pylori (Table 3)
all of which have been implicated as risk factors in Alzheimer’s disease (23;24)
. Indeed, in infected Alzheimer’s disease patients, H.Pylori eradication can
have beneficial effects on cognition (25) . C.Pneomoniae (penta- and hexapeptides)
B.Burgdorferri (pentapeptides) and H.Pylori (Penta- and hexapeptides) proteins
all contain matching beta-amyloid fragments. Tooth loss and periodontitis
have also been cited as risk factors in Alzheimer’s disease (26;27) . Porphyromonas
gingivalis and Streptococcus mutans are major cause of periodontitis, and
proteins from these bacteria also contain the VGGVV and other internal beta-amyloid
sequences (Table 3).
There are two case reports in the literature recording
subjects diagnosed with Alzheimer’s disease, both with a three year history
of dementia. Cryptococcal meningitis was subsequently diagnosed, and in both
cases, antifungal treatment resulted in an almost complete recovery (28;29)
C.Neoformans is the agent responsible for Cryptococcal meningitis and contains
a number of proteins showing a striking similarity with beta-amyloid internal
sequences, including exact homology with an N-terminus octapeptide and a c-terminus
heptapeptide as well as the VGGVV sequence (Table 3).
Allergenic proteins
A search of the allergen databases showed that stretches within the beta-amyloid
peptide, exactly match those in several common allergens, particularly in
European (hexapeptide) and American (pentapeptide) house dust mites (Table
2, Fig 3 ) and in a rice fungus Aspergillus Oryzae. Rather less pentapeptide
or more matches were found in this class but very many common pollen, fungal
, insect venom, domestic animal (cat, cattle and horse) and food allergens
(beans, rice, potato, tomato and nuts ) contain tetrapeptide sequences matching
those of beta amyloid . House dust mites, as well as containing relatively
large consensual sequences, display a large number of smaller peptide matches
along the length of the beta-amyloid peptide (Table 2) suggesting that these
ubiquitous and unavoidable creatures could play a rather unexpected role in
Alzheimer’s disease.
Immune activation in the Alzheimer’s disease brain
A number of immune-system related proteins are found in amyloid
plaques or neurofibrillary tangles. Interleukin 1 alpha, interleukin 6, and
tumor necrosis factor are all been localised within plaques (30) and acute
phase proteins involved in inflammation, such as amyloid P, alpha-1 antichymotrypsin
and C-reactive protein are also plaque components (31) while Immunoglobulin
G is located in the plaque corona (32) . Large increases in IgG levels have
been recorded in the brain parenchyma, in apoptotic dying neurones and in
cerebral blood vessels in the Alzheimer’s disease brain (33) . Complement
component C3 is found in Alzheimer’s disease amyloid plaques along with complement
C4 (34) . Complement components Clq, C3d, and C4d are present in plaques,
dystrophic neurites and neurofibrillary tangles (5) .
The membrane attack complex (MAC), composed of complement
proteins C5 to C9, forms a channel that is inserted into the membranes of
pathogens, destroying them by lysis. These components cannot be detected in
temporal cortex amyloid plaques in Alzheimer’s disease (34;35) . However the MAC complex is present in dystrophic neurites
and neurofibrillary tangles (5) and others have detected this complex in neuritic plaques
and tangles, along with deposition of C1q, C3 and clusterin
(36) . The membrane attack complex
has also been detected in the neuronal cytoplasm in AD brains and associated
with neurofibrillary tangles and lysosomes (4) .The presence of the MAC complex in neurones might suggest
that neuronal lysis by the MAC complex could contribute to neuronal cell death
(5) .
Pathogen seropositivity in Alzheimer’s disease
Increased seropositivity to IgM anti HSV-1 antibodies has been
reported to predict the risk of subsequently developing Alzheimer’s disease
(14) . Increased seropositivity in Alzheimer’s disease has also been reported
for Helicobacter pylori (37) and increased HHV-6 immunoreactivity has been
observed in Alzheimer’s disease CSF samples (38) . No differences in the seropositivity
of Adenovirus, Chlamydia Group B, Coxiella burnettii, Cytomegalovirus, Herpes
simplex virus, Influenza A, Influenza B, Measles and Mycoplasma pneumoniae
were found in a study of 33 Alzheimer’s disease patients and 28 controls (39)
. However, there are a large number of diverse pathogenic antigens with homology
to beta-amyloid, any of which may have been present, potentially provoking
immune response-related neuronal loss, many years prior to seropositivity
testing.
Antibodies and antigenicity
The tenet of the autoimmune hypothesis is that viral and other
pathogens and environmental allergens with homology to beta-amyloid will trigger
the production of antibodies that also target the beta-amyloid peptide. Those
provoking a robust immune, inflammatory and complement response risk killing
beta-amyloid containing neurones. Antibodies to the Potato virus (15) and
to Borrelia antigens (24) ) have already been shown to label amyloid plaques
in the Alzheimer’s disease brain and antibodies to a phage epitope (AEFRH)
also label beta-amyloid (40) .
Potential cross-reactivity can partly be tested in silico
but ultimately requires the characterisation of the autoimmune beta-amyloid
epitopes present in Alzheimer’s disease and cross-reactivity testing between
pathogenic and environmental antigens and beta-amyloid. Thus far, the precise
epitopes labelled by beta-amyloid autoantibodies have not been fully characterised.
The initial rendezvous of antigens with the immune system is
with B-cells which bind to, engulf and digest the antigen. The B-cell epitope
antigenicity prediction (20) for beta-amyloid is illustrated in Figs 2 and
3, over which are laid the pathogens and allergens that match particular sequences
within the beta-amyloid peptide. As can be seen, Coxsackie, HIV-1, HSV-1,
Hepatitis C, influenza, mumps, the respiratory syncitial virus, rhinoviruses
(common cold) and clostridia, enterobacteria and vibrio phages match sequences
that lie within predicted antigenic regions of beta-amyloid (Fig 2). An increase
in immunogenicity is also observed within the VGGVV sequence, which has been
used as an epitope to label beta-amyloid (7) . Allergenic proteins whose sequences
match those of antigenic portions of beta-amyloid include those from fungal,
nut, pollen, insect venom and house dust mites (Table 2, Fig 2).
As a further theoretical test, all consecutive tetrapeptide sequences
within beta-amyloid were screened against the human epitope and the HIV-1
molecular Immunology databases to determine which antigens contain these epitopes.
As shown in Tables 2 to 7 many viral, fungal, bacterial, parasite and allergen
antibodies contain tetrapeptide sequences from the beta-amyloid peptide. In
addition, apart from beta-amyloid isoforms and a small number of mammalian
proteins, the major matching epitopes were concentrated in viral, bacterial,
fungal and allergen classes (i.e. not other species or other mammalian proteins).
These databases contain over 73,000 epitopes, so again the results returned
are highly specific to these classes. This at least shows that antibodies
that have been used to label these viruses, pathogens and allergens, contain
beta-amyloid sequences, and that potential cross-reacting antibodies are concentrated
in these classes.
The ability of autoantibodies to adsorb beta-amyloid can be considered
as useful, and certain of these appear to be associated with reduced plaque
volume (1) . Other autoantibodies, present in normal and Alzheimer’s disease
sera, also possess catalytic properties and are capable of destroying the
toxic peptide. Such antibodies do not form stable immune complexes and are
less likely to activate immune inflammatory and complement related cell lysis
(2) . This is where the danger lies, as antibodies capable of mounting a full-blown
immune response against beta-amyloid are likely to kill the cells in which
the peptide resides. It is relatively common to find Alzheimer’s disease pathology
at autopsy (plaques and tangles) in patients who were cognitively normal shortly
before death (41) . In such patients, it is tempting to suggest that immune-activating
antibodies are rare.
Catalytic beta-amyloid auto-antibodies cleave the beta-amyloid
peptide primarily between the two histidines (H/Q) at Abeta13-14
, and to a lesser extent at other positions as shown in Figs 2 and 3 {Taguchi,
Planque, et al. 2008 1720 /id} . The major cleavage site is within a non-immunogenic
region of the peptide. Interestingly, several plant viruses (Amaranthus, Pepper,
Pistachio, Potato, Rice, Soybean, Sunflower and Zucchini (courgette) and allergens
(Rice, Pistaccio, Soybean as well as the cat allergen and insect venoms)
express proteins homologous to this particular region. Rather intriguingly,
several of these plants/vegetables are constituents of the Mediterranean diet
which has been reported to reduce the risk of developing Alzheimer’s disease
(42) . Such diets impact on cholesterol/lipoprotein function and on atherosclerosis,
a major contributory factor in Alzheimer’s disease (43;44) but could also
conceivably be related to beneficial antibodies generated by the ingestion
of common plant viruses, a possibility that also suggests potential immunisation
strategies.
Certain proteins from three cancer-causing viruses, Epstein-Barr
(HSV-4), Hepatitis B and the human Papillomavirus virus are also homologous
to this catalytic antibody target region, suggesting a plausible explanation
for the inverse association between cancer and Alzheimer’s disease (45)
Vaccinations and Alzheimer’s disease
It has been noted that the risk of developing Alzheimer’s disease
was reduced following vaccination against diphtheria, influenza, polio or
tetanus (46) . Again, sequences within proteins of these pathogens are homologous
to beta-amyloid as shown in Fig 2, (which represents the viral proteins rather
than the epitopes used for vaccination). This is rather encouraging as it
suggests that, if the effect of vaccination is due to beta-amyloid homology,
other vaccines might similarly reduce the incidence of Alzheimer’s disease.
The papillomavirus vaccine, already used to prevent cervical cancer (47) ,
is a prime candidate as a viral protein matches the beta-amyloid region targeted
by catalytic autoantibodies. However, as there is an evident danger of creating
potentially toxic beta-amyloid autoantibodies, characterisation of the epitopes
targeted by such vaccines must be of prime concern.
Epidemiological studies
These observations suggest that Alzheimer’s disease may have
an autoimmune component, triggered by antigenic proteins with homology to
beta-amyloid. Such a scenario helps to explain several, indeed most, epidemiological
features of Alzheimer’s disease. For example, Atopy and autoimmune diseases
are common in Alzheimer’s disease (48;49) , and could relate to allergen
homology with beta-amyloid. Alzheimer’s disease prevalence is higher in women
and in Afro-Americans (50) , as is HSV-2 prevalence (51) .
. Herpes simplex infection is a risk factor in Alzheimer’s disease
acting in synergy with the APOE4 allele (52) and dementia, with Alzheimer’s
disease pathology is common in HIV-1 infected patients (53) , a situation
perhaps explained by viral protein homology with beta-amyloid. Such homology
with pathogen proteins could also explain the association of Borrelia Burgdorferri,
Helicobacter pylori, C.Pneumoniae (23;24) and by inference P.Gingivalis and
Streptococcus mutans (oral pathogens causing periodontitis and tooth loss
(26;27) ) with Alzheimer’s disease. The incidence of Alzheimer’s disease
is also related to the number of pregnancies (54) , which might be explained
by greater exposure to common childhood illnesses (cf phage/bacterial homology
with beta-amyloid). It has also been noted that nuns, who are less exposed
to sexually transmitted pathogens, show some resistance to the ravages of
Alzheimer’s disease (41) . An autoimmune scenario may also explain why the
use of non-steroidal anti-inflammatories reduces the risk of developing Alzheimer’s
disease (55) as do fish consumption and the Mediterranean diet (42;56) .Such
diets are rich in N-3 polyunsaturated fatty acids, which also possess anti-inflammatory
properties (57) . The beneficial effects of these diets and also of statins
(58) are likely to be related to multiple problems in cholesterol and lipoprotein
homoeostasis in Alzheimer’s disease (59) but may also be related to pathogens
and the immune system. For example viral entry is often lipid dependent a
a factor related to fusion of the viral lipid envelope with cell membranes,
and the cellular entry of HIV-1 and herpes simplex is cholesterol and lipid
raft -dependent and blocked by nystatin (60;61) . Statins also habe immunosuppressant
properties (62) . The potential role of viruses as constituents of the Mediterranean
diet has been discussed above in relation to their homology with beta-amyloid
regions targeted by beneficial catalytic autoantibodies. The inverse association
between cancer and Alzheimer’s disease might also be explained in these terms
(see above) and the beneficial effects of vaccination can also be related
to pathogen homology with beta-amyloid (diphtheria, influenza, tetanus and
polio). The risk promoting effects of aluminium in Alzheimer’s disease (63)
might also be explained by its common self-prescribed use as an antacid in
gastrointestinal disturbances (64) often caused by phage/bacterial pathogens
with homology to beta-amyloid.
In short almost all epidemiological observations related to Alzheimer’s
disease can be explained in terms of pathogens and/or autoimmunity.
Late-onset Alzheimer’s disease susceptibility genes
Four genes, Apolipoprotein E, clusterin, complement receptor 1 and PICALM,
are the main suspects in Alzheimer’s disease (65-67) each of which can be
implicated in viral life cycles. For example, APOE binds to HSV-1 (68) and
Hepatitis C viruses (69) and the Alzheimer’s disease risk allele, APOE4, is
also related to HSV-1 and HIV-1 infection, although, curiously, it protects
against Hepatitis C infection (70) . Complement receptor 1 and clusterin
are both involved in the complement cascades that play a crucial role in pathogen
defence (71) . In addition the influenza virus and HSV-1 both bind to complement
receptor 1 on erythrocytes (72) . One of the receptors used by
Herpes simplex for cellular entry is the Mannose-6-phosphate receptor, M6PR
(73) . This receptor binds to clusterin (74) and its traffic through the endosomal compartments is controlled
by PICALM, whose overexpression reduces M6PR localisation in endosomes, suggesting
blockade of its transport from the plasma membrane or the trans-golgi network
(75) . The Herpes simplex and Influenza viruses also uses exportin
(Crm1) dependent pathways for nuclear egress (76;77) . PICALM and other endocytic-regulatory proteins bind to Crm1
(78) .
Many minor genes (see www.polygenicpathways.co.uk/alzpolys
for references) are also implicated in viral life cycles. For example lipoprotein
receptors implicated in Alzheimer’s disease (LRP1, LDLR, VLDLR) are used by
human rhinoviruses to gain cellular entry (79) . The nectin receptor PVRL2
is a Herpes viral receptor (80) , as is insulin degrading enzyme (81) . In
addition over 30 immune related genes (chemokines, cytokines, complement related,
toll receptors HLA-antigens) have been implicated as risk factors. All of
these might be expected to modulate immune defence.
Given the plethora of viral and environmental beta-amyloid homologues,
and the presence of autoantibodies to beta-amyloid in healthy aged subjects,
it seems likely that the functional gene variants in the control group, rather
than the Alzheimer’s disease risk promoting genes, may be more important.
These are presumably those that prevent the ravages of autoimmunity.
Familial Alzheimer’s disease
Familial early-onset Alzheimer’s disease is caused by a number
of different mutations in presenilins and the APP gene, including several
mutations as APP717 (V/I, V/M, V/G, V/L) (London mutation and
others) and at KM670/671NL (Swedish mutation) (inter alia :see http://www.molgen.ua.ac.be/ADMutations/)
(82) . These are not within the beta-amyloid sequence. Both mutations modify
APP processing and increase the generation of Abeta 1-42 (83;84)
. In so doing, they may thus be able to increase the probability of Abeta
encountering autoantibodies, both in the brain and in the periphery resulting
in a feed forward further generation of autoantibodies by the immune system.
The APP717 mutant is within two gamma-secretase cleavage
sites generating an undecapeptide as shown in Fig 1. The Swedish mutation
is immediately prior to the beta-secretase cleavage site that, with gamma-secretase,
generates beta-amyloid and frees the native or mutant APP670/671
terminus (85) . Homology searches against viral and bacterial proteomes were
performed using the native and mutant forms of the APP717 undecapeptide and the
nonapaptide upstream of the Swedish mutation sites (Fig 1). The APP717G
mutant increased the predicted B-type immunogenicity of the peptide,
but other mutants were without effect (not shown).
Native APP717 is homologous to several phages, viruses
and bacteria, many of which are common; for example Enterobacteria, lactococcus
and staphylococcus phages, adenovirus, cytomegalovirus, the parainfluenza
virus and rotavirus and the less common rabies and coronaviruses (Table 4).
This native form also shows homology with several bacterial species, which
for the most part, with the exception of Lactobacilli, are pathological rather
than commensal (Table 5).
The various APP717 mutations alter this matching profile
in a number of ways. For example the V/F mutation creates matching peptides
to Herpes viruses HSV-2, 3 and 8 and markedly increases the number of hits
in relation to homology, particularly within bacterial flora (mainly due
do many strains of the commensal E.Faecalis) (Tables 4 and 5).
The APP717 V/G mutant creates a peptide region homologous
to proteins from human herpes virus 6, a pathogen with seroprevalence approaching
100% (86) and to the JC and BK polyomaviruses which also display high seroprevalence
in the normal population ( 39% and 82% respectively (87) ) . This mutant also
creates regions homologous to the endemic soil bacterium B.Cereus, to a bacterium
causing verrucas, and to P.Gingivalis a constituent of the oral flora that
plays a role in periodontitis and tooth loss (tables 4 and 5).
The V/I mutant creates homologues to proteins from over 30 Rhinococcal
strains causing the common cold, to the mumps virus and to the common Norovirus
responsible for vomiting sickness, as well as to the endemic soil bacteria
B.Cereus and S.Aureus, and the commensal E.Coli and P.Gingivalis (Table 4,5).
The V/L mutant creates regions homologous to proteins from Influenza A and
B viruses and to Hepatitis C and Herpes viruses 4 and 5, as well as to the
commensal E.Coli and Streptococcus Hominis (Tabke 4,5).
The native APP670/671 peptide is homologous to a number
of influenza viruses, which are almost exclusively avine. The Swedish mutant
increase the number of viral matches to the upstream peptide, particularly
to enterobacterial phages, and creates homologous regions to several species
of human adenoviruses, which also show high seroprevalence (88) , and to the
respiratory syncitial virus as well as to human rhinoviruses and again to
E.Coli. Several of these mutant peptides are homologous to mycobacterial phages
(see Tables 6,7 ), perhaps explaining why tuberculosis (as well as possession
of the HLA-DR3 allele) have been reported as risk factors in familial
Alzheimer’s disease (49) . The epitopes generated by these mutants are
again concentrated within viral and pathogen proteins, particularly the Vaccinia
Virus (Tables 4-7). There do not appear to have been any studies on autoantibodies
in familial Alzheimer’s disease but the large number of viral and bacterial
peptides perfectly matching the mutant surrounds suggests that autoimmunity
may also play a major role in this disorder. Although clearly genetic, its
pathology may be autoimmune related, as well as to, or perhaps rather than
aberrant beta-amyloid processing.
In short, these mutants, in different ways, all increase the
number and variety of viral and bacterial matches, to commensal (Particularly
E.Coli, E.Faecalis and P.Gingivalis ) as well as to pathological species (Clostridia,
Mycobacteria, Vibrio, inter alia) and create homologous regions to
very common pathogens causing the common cold, influenza, herpes infections
and periodontitis.
APP transgenic mice
If autoimmunity rather than problems in APP processing is relevant,
this could explain why APP transgenic models do not faithfully mimic Alzheimer’s
disease (i.e. extensive cholinergic neuronal loss, loss of hippocampal afferents
and efferents and massive cortical degeneration) (89) . There have been two
studies assessing the effects of infection in APP transgenic mice. Repeated
Streptococcus Pneumoniae infection had no effect on pathology or behaviour
in APP Tg2576 transgenic mice (Swedish mutant) (90) . S.Pneumoniae displays
homology to both the native and mutant forms of the relevant peptide (Table).
Borna virus infection resulted in a reduction in beta-amyloid immunoreactivity
in the brains of infected Tg2576 transgenic mice (91) . This virus also displays
homology to both the native and mutant forms of the peptide (Table). S.Pneumoniae
also expresses proteins containing the VGGVV sequence, as do a large number
of other bacteria (not shown), while a homology search for Borna virus proteins
containing VGGVV yielded no hits. However, in relation to autoimmunity, it
is not the infection itself, but the antibodies generated in response to the
infection that could cause the problem. A more suitable test might be repeated
challenge with specific antigens in APP transgenic models.
Other models capable of producing cerebral beta-amyloid deposition
in mice include C.Pneumoniae (92) or HSV-1 infection (11) , the latter also
producing hippocampal and entorhinal cortex neuronal loss and memory deficits,
perhaps more faithfully reproducing the pathology of Alzheimer’s disease
(13) . Such models may be useful in APP transgenic mice.
Beta-amyloid and other Vaccines and therapies.
.
The F,G and I APP717 mutants all generate peptides
homologous to the related Vaccinia and Variola (Smallpox) viruses (Table 4,6)
. While vaccination against diphtheria, flu, tetanus and polio have been reported
to reduce the risk of late-onset Alzheimer’s disease (see above) there is
also a possibility of vaccine cross-reactivity with endogenous peptides such
as beta-amyloid. Many of the epitopes formed by these mutant peptides are
contained within Vaccinia viral proteins. There is thus a possibility that
smallpox vaccination may be a contributory factor in familial Alzheimer’s
disease, although this requires characterisation of the epitopes present in
the vaccine, which was generated from the virus rather than from peptide components.
The vaccinia virus has also been used as a primary vaccination against smallpox
(93) following on from Edward Jenner’s pioneering experiments with cowpox
over 200 years ago (94) . This is a contentious area, but the possibility
that vaccination might trigger nefarious autoantibody generation does need
to be addressed, particularly in future studies.
The potential use of beta-amyloid antibodies is based on their
ability to reduce plaque burden and neurite dystrophy in APP transgenic mice
(95)
Several studies have demonstrated that beta-amyloid antibodies reduce plaque
burden in APP transgenic models and that they can also improve cognitive performance
(96) . However amyloid antibodies extracted from the serum of old APP transgenic
mice potentiate the toxicity of beta-amyloid and Alzheimer’s disease patients
display an enhanced immune response to the peptide (97) . Again in transgenic
mice, different immune backgrounds can influence the type of immune responses
elicited by beta-amyloid. For example B-and T-cell responses to beta-amyloid
can be modified in HLA-DR3,
-DR4,
-DQ6 or -DQ8 transgenic mice (98)
. HLA-antigen diversity in Man is also likely to determine the outcome of
beta-amyloid/antibody interactions.
The results of this survey suggest that beta-amyloid autoantibodies
might cause as well as mitigate against Alzheimer’s disease pathology. Beta-amyloid
vaccination in Alzheimer’s disease (against Abeta1-42) has so far
not been successful and sadly resulted in meningoencephalitis and the death
of a patient (99) . While certain beta-amyloid antibodies may reduce plaque
burden, there is an evident risk that they may also trigger an auto-immune
response, potentially killing beta-amyloid containing neurones. Catalytic
autoantibodies are less able to form stable immune complexes and likely represent
the safest way forward in this area (2) . It is intriguing that the beta-amyloid
region targeted by catalytic autoantibodies matches peptide sequences of viruses
that are constituents of the Mediterranean diet and of cancer-inducing viruses
(Epstein-Barr, Hepatitis B, and the papillomavirus) as both cancer and the
Mediterranean diet are inversely associated with Alzheimer’s disease risk
(see above). Vaccines to the human papillomavirus already exist (47) and could
perhaps be considered as a therapeutic or preventive option in Alzheimer’s
disease, after due consideration of the epitope matches.
If beta-amyloid autoantibodies are the culprits, techniques such
as immunoadsorption, which has proved to be of benefit in Myasthenia gravis
(autoimmunity to acetylcholine receptors (100) or immunosuppressant use might
be considered as potential therapeutic options. There is indirect evidence
in support of such treatment. Natural immunoadsorbants include silica, which
is however toxic, (101) , tryptophan and phenylalanine (102) . Levels of silica
in drinking water are inversely related to Alzheimer’s disease risk (103)
and serum tryptophan levels are markedly depleted in Alzheimer’s disease patients,
a marker of immune activation (104) .Phenylalanine plasma levels are in contrast
increased in Alzheimer’s disease (105) . Fish oil (see above) also suppresses
T-Lymphocyte activation (106) and statins, which have also been reported to
reduce Alzheimer’s disease risk (58) are also immunosuppressant (62) .
Heterogeneity in genetic and epidemiological studies
If there is one factor common to polygenic disorder research
it is the discordance of genetic and environmental association data in this
and other diseases. However, if, as suggested by this survey, there are dozens
of potential Alzheimer’s disease triggers, all funnelling towards a common
cause, this heterogeneity becomes part of the answer and not part of the problem.
Different gene products are related not only to human physiology, but also
to pathogen life cycles (e.g. rhinoviruses and lipoprotein receptors, APOE
and Herpes Simplex or influenza and complement receptor 1) , a situation that
has been discussed in relation to Alzheimer’s disease and Schizophrenia susceptibility
genes and pathogens implicated in these disorders (107;108) . Viruses and
bacteria are not uniformly distributed worldwide, nor is antibody seroprevalence,
and different environmental risk factors may similarly vary from region to
region. The heterogeneity observed in these studies thus has a rational basis,
reflecting the heterogeneity of cause, and need not necessarily be considered
as a statistical artefact
Relevance to other autoimmune and genetic diseases and evolutionary aspects
.
A homology search against viral proteomes with known autoantigens
from multiple sclerosis, myaesthenia gravis, pemphigus vulgaris, systemic
lupus erythematosus, and Chronic obstructive pulmonary disease retuned viral
matches, which in all cases were relevant to the viruses implicated as risk
factors in each disease (Table 8) , also identifying other suspects. Furthermore,
in other genetic disorders, Huntingtons’s disease and
other polyglutamine repeat disorders (Dentatorubropallidoluysian atrophy,
Kennedy disease and Spinocerebellar ataxias (109;110) ) and Cystic fibrosis,
the mutations created homologues with several viral or phage proteins. The
QQQ polyglutamine triplet is a B-cell epitope according to the BEpiPred server
and each successive QQQ addition increases the overall antigenicity of the
resulting peptide (not shown). As an example of a risk promoting gene in polygenic
disorders, APOE4 also aligns with several relevant viral proteins. Given the
capacity of the immune system to generate vast repertoires of antibodies,
some of these pathogens will no doubt generate antibodies that also target
important human proteins implicated in autoimmune and genetic diseases. The
genetic mutations often create physiological problems related to their normal
function and the effects of the mutation, but the overall effect of the mutation,
particularly in relation to autoimmune-related degeneration, may be compounded
by these viral matches.
Phages and viruses are the simplest form of “life”, as defined
by the possession of DNA/RNA and a proteinaceous structure, and were long
ago proposed as the origin of higher cellular organisms (111;112) . There
are currently 2463 viral genomes in the NCBI database, probably representing
but a small proportion of those existing on the planet. While perhaps responsible
for our origin these predecessors may have donated a legacy of human viral-derived
proteins that closely match antigenic proteins in the currently existing virome
that could be responsible for many human diseases including genetic disorders,
autoimmune disorders, polygenic diseases, and those that have so far baffled
research, such as ME (Myalgic encephalomyelitis/chronic fatigue syndrome) and fibromyalgia.
This evidently has enormous implications for therapy and prevention, not only
of autoimmune disorders, but also of polygenic and human genetic disorders,
until now generally regarded as unassailable. Vaccination, using epitopes
against the non homologous human protein regions of the phages and viruses
could perhaps destroy the pathogen and prevent the associated problems related
to autoimmunity.
Summary and conclusions
Many common viruses, phages, bacteria, fungi, parasites and allergens
express proteins with marked homology to antigenic and other regions of the
beta-amyloid peptide. Epitope similarity with beta-amyloid is concentrated
within these classes and it seems likely that these antigens provide a source
of the beta-amyloid autoantibodies found in the ageing population and in Alzheimer’s
disease. While some of these antibodies may be benevolent, others may stimulate
immune, inflammatory and other defensive measures, including complement mediated
cell lysis that could kill the neurones in which the peptide resides. Activation
of the immune system is supported by the presence of many immune-related proteins
in Alzheimer’s disease amyloid plaques, and by the presence of the complement
membrane attack complex in Alzheimer’s disease neurones. Reduced serum tryptophan
levels, and an increased immune response to beta-amyloid also suggest immune
activation. Familial Alzheimer’s disease may also have a strong autoimmune
component, as the various APP mutants convert the surrounding peptides to
matches to commensal bacteria (E.Coli, E.Faecalis and P.Gingivalis) and to
viruses with a high seroprevalence (HHV-6, polyoma viruses, influenza and
the common cold rhinoviruses). The categorisation of Alzheimer’s disease as
an autoimmune disorder explains most of the epidemiological observations in
Alzheimer’s disease and the major genes implicated in Alzheimer’s disease
are all related to viral life cycles and/or to the complement arm of the immune
defence network.
Diseases caused by these viruses, fungi or the bacteria infected
with the phage/beta-amyloid homologues are very common and often recurrent
(e.g. colds, influenza gastroenteritis or food poisoning) as is exposure to
certain allergens (e.g. dust mites , cat, cow, horse allergens, pollen and
food allergens, insect stings, marine algae etc) Over time, and with increasing
age, the major risk factor in Alzheimer’s disease (113) , antibodies that
may also target beta-amyloid are more likely to be produced, gradually increasing
the probability of a self-immune attack on neurones containing the beta-amyloid
peptide. There may thus be dozens, if not hundreds of triggers promoting Alzheimer’s
disease risk, all funnelling towards an autoimmune scenario. If this is the
case, then Alzheimer’s disease might be considered as an autoimmune disorder
and immunosuppressants or antibody adsorption might have a role to play in
its therapy, once diagnosed. C.Neoformans eradication can result in a complete
recovery, in very rare cases of diagnosed dementia (28;29) , and Helicobacter
elimination has been reported to ameliorate cognitive function in infected
Alzheimer’s disease patients (25) . Aggressive targeting of opportunistic
pathogens capable of mimicking the beta-amyloid peptide might thus be considered
as a therapeutic option. Because so many are homologous to beta-amyloid, such
therapy might well have to be tailored to individual pathogens, depending
on the species identified by serum assay.
Vaccination against common diseases has already been shown to
reduce the risk of developing Alzheimer’s disease (46) and, in the long term,
vaccination against other common viruses and bacteria might also be of benefit,
although it is evident that potential vaccine antibody cross-reactivity with
beta-amyloid must be a prime concern. One such vaccine may already exist in
the form of that for the human papillomavirus.
. In summary, the close homology of diverse viral, fungal bacterial
and allergenic antigens with beta-amyloid and to peptides generated by APP
mutations suggests an autoimmune component to familial and late-onset Alzheimer’s
disease, triggered by these antigenic homologues. The autoimmune scenario
explains many epidemiological observations and genetic studies also implicate
the immune system and viral life-cycles. If autoimmunity is important, vaccination,
viral and pathogen elimination and immunosuppressant therapy might be expected
to play a role in the prevention and therapy of Alzheimer’s disease and perhaps
provide new rationales for developing a cure. This type of viral matching
is also relevant to other autoimmune and genetic disorders and may be a near
universal phenomenon reflecting our viral ancestral roots. This generality
could open new therapeutic avenues in many human diseases.
Acknowledgements: Thanks to the numerous authors for reprints, to Oliver
Chao and Nasire Mahmudi for finding others, and to Maria Jesus Martin at Uniprot
and Tao Tao at NCBI for help with the mysteries of BLAST and Clustal alignment
settings.
Table 4: The
effects of the APP717 mutations (in Red) on homology to viral proteins.
Protein accession numbers are provided and amino acid matches are indicated
by the asterisks or by the red letter of the mutant amino acid. Species with
proteins containing epitopes matching those of the peptide amino acids are
also shown (Marked by E). Phages infecting commensal bacteria and common viruses
(eg Rhinoviruses and influenza) are highlighted in bold.
Native |
I |
A |
T |
V |
I |
V |
I |
T |
L |
V |
| Schistosoma
japonicum
Bovine papillomavirus - 4
Protein tyrosine phosphatase receptor Homo sapiens |
|
|
Epi |
E |
E |
E |
|
|
|
|
| Vaccinia virus: Nicotinic receptor Homo
sapiens |
|
|
|
Epi |
E |
E |
E |
|
|
|
| Entamoeba histolytica
Lymphocytic Choriomeningitis virus
Dengue virus 2
Nicotinic receptor Homo sapiens |
|
|
|
|
Epi |
E |
E |
E |
|
|
Bacillus Phage NP_046589
Rabies virus ACN38519
Salmonella Phage YP_001742070
Sendai Virus BAC79139 |
* |
* |
* |
* |
* |
|
|
|
|
|
Aeromonas Phage YP_238915
Enterobacteria phage AC14704
Human Coronavirus 229E CAA49377
Mossman Virus NP_958055
Pseudomonas phage YP_418190
SARS Coronavirus ACZ71766 |
|
|
* |
* |
* |
* |
* |
|
|
|
Staphylococcus phage
YP_024500 |
|
|
|
* |
* |
|
* |
* |
* |
* |
Aeromonas phage NP_932517,
Acanthamoeba polyphaga mimivirus
AAV50741
Flavobacterium Phage YP_112527
Mycobacterium Phage YP_002242149
Salmonella phages: Pseudocowpox ADC53802
Uncultured Phage
ADF97555
Paris Polyvirus X ABF74755 |
|
* |
* |
* |
* |
* |
|
|
|
|
Lactococcus Phage P_002875673
Pseudomonas Phage YP_003422512
Ralstonia Phage YP_001165297
Newcastle Disease virus ACZ72939 |
|
|
|
|
* |
* |
* |
* |
* |
|
Human herpesvirus 5 (Cytomegalovirus) AAS48926
|
|
|
|
* |
* |
* |
* |
* |
|
|
SARS coronavirus AAY60778
Burkholderia phage YP_001111213
Enterobacteria phageYP_001837048
Human rotavirus A BAF95721
Human parainfluenza virus ACZ95446
Human adenovirus 1 AP_000521
Lactate dehydrogenase-elevating virus NP_042576 |
|
|
|
|
|
* |
* |
* |
* |
* |
Vaccinia virus |
|
|
Epi |
E |
E |
F |
|
|
|
|
Vaccinia virus |
|
|
|
Epi |
E |
F |
E |
E |
|
|
Vaccinia virus Plasmodium Falciparum |
|
|
|
|
Epi |
F |
E |
E |
|
|
Apis Mellifera |
|
|
|
|
|
F |
Epi |
E |
E |
|
Human Herpes Virus 2 AAU93410 |
|
|
* |
* |
* |
F |
* |
* |
|
|
| Ralstonia
Phage YP_001949949
Streptococcus PhageCAA87730 |
|
* |
* |
* |
* |
F |
|
|
|
|
Bacillus phage NP_046680 |
|
|
* |
* |
* |
F |
* |
|
|
|
| Human papilloma
virus BAC79139 |
|
|
|
|
* |
F |
* |
* |
* |
|
| Human herpesvirus
3 (Varicella Zoster) ACL67882
Human herpesvirus 8 AAW31674
Verbena virus Y YP_001936181
Vaccinia virus CAM58311
Variola virus ABF23093 Mycobacteriophage ADB93732
Enterobacteria phage NP_861932
Bacteriophage phi-105 BAA36658 |
|
|
|
|
|
F |
* |
* |
* |
* |
Bacillus phage SPBc2 NP_046680 |
|
|
* |
* |
* |
F |
* |
|
|
|
Streptococcus phage PH10 YP_002925182
Enterococcus Phage YP_003358819
Feldmannia irregularis virus AAR26941
Staphylococcus prophage NP_061652
Human Papilloma virus Type 116
YP_003084352
and 103 YP_656498 |
|
|
|
|
* |
F |
* |
* |
* |
|
Verbena Virus Y YP_001936181 |
|
|
|
* |
|
F |
* |
* |
* |
* |
SARS coronavirus |
|
|
|
Epi |
E |
G |
E |
|
|
|
| C.Tetani
Human metapneumovirus
Human papillomavirus – 1 |
|
|
|
|
Epi |
G |
E |
E |
|
|
| Deerpox virus,Guanarito virus, Hepatitis
A virus, Human herpesvirus 5
Human T-cell lymphotrophic virus type 1
Junin virus, Lymphocytic choriomeningitis virus
Mycobacterium tuberculosis, SARS coronavirus
Vaccinia virus WR, Vibrio sp. |
|
|
|
|
|
G |
Epi |
E |
E |
|
Vibrio Phage NP_899405 |
|
|
* |
* |
* |
G |
* |
* |
|
|
Puumala virus ACJ22524
|
|
|
|
* |
* |
G |
* |
* |
* |
|
BK Polyomavirus ADA67892 |
|
|
|
* |
* |
G |
* |
* |
|
|
Bacillus phage SPO1 YP_002300316
Lymphocyti choriomeningitis virus ABI96825
Xanthomonas phage YP_001469625 |
|
|
|
|
|
G |
* |
* |
* |
* |
Bacillus phage G |
|
* |
* |
* |
* |
G |
* |
|
|
|
| Staphylococcus prophage NP_061629
JC polyomavirus BAB93084 |
|
|
|
|
* |
G |
* |
* |
* |
|
Highlands J virus ACZ34298
JC Polyoma virus BAC66416 |
|
|
* |
* |
* |
G |
* |
|
|
|
Amapari virus YP_001649216
Dandenong virus ABY20732,
Guanarito virus AAP44540
Human T-lymphotropic virus AAT38829
Junin virus AAT40446
Latino virus YP_001936026
Lujo virus
YP_002929493
Lymphocytic choriomeningitis virus ABB88931
Oliveros virus YP_001649214
Parana virus YP_001936028
Pirital virus
AAP44541
Vaccinia Virus ABZ80096
Variola virus ABF26311 |
|
|
|
|
|
G |
* |
* |
* |
* |
Aeromonas phage NP_944088
Bacillus phage SPOI YP_002300316
Burkholderia phage NP_944301
Iodobacteriophage YP_002128470
Mycobacterium Phage YP_001994770
Vibrio Phage AAN74000
Xanthomonas phage YP_001469625 |
|
|
|
|
|
G |
* |
* |
* |
* |
Lactobacillus phage ADA79910
BK polyomavirus ADA67892
Mycobacterium phage ACU41960 |
|
|
|
* |
* |
G |
* |
* |
|
|
Human herpesvirus 6 AAA16727
Human herpesvirus 6B BAA78241
Enterobacteria phage RB69 NP_861857 |
|
|
* |
* |
* |
G |
* |
|
|
|
| Dengue virus 3 HSP60 Homo
Myelin P2 Homo |
|
|
Epi |
E |
E |
I |
|
|
|
|
| SARS Corona virus
Vaccinia virus |
|
|
|
Epi |
E |
I |
E |
|
|
|
| Vaccinia virus WR
Plasmodium falciparum
Lymphocytic choriomeningitis virus
Nicotinic receptor Homo sapiens |
|
|
|
|
Epi |
I |
E |
E |
|
|
Rhinovirus 50 ACK37391 and >30 other Rhinovirus strains |
* |
|
|
* |
* |
I |
|
* |
* |
* |
| Rhino virus 38 ABF51189
Virus PK224 ADF80719
Aeromonas phage NP_943894
Enterobacteria phage SP ACY07251
Enterobacteria phage F1 NP_695026 (and others)
Listeria phage YP_002261439
Acanthamoeba polyphagamimivirus YP_001427182
Human
herpesvirus 4 YP_401711
Human herpesvirus 5 ACS91947
Aeromonas phage NP_943894
Lymphocystis disease virus NP_078753
SARS coronavirus AAU93319 |
|
|
* |
* |
* |
I |
* |
|
|
|
Vaccinia Virus NP_048189 |
|
|
|
* |
* |
I |
* |
* |
* |
* |
| Staphylococcus phageYP_238539
Vibrio Phage NP_899546 |
|
* |
* |
* |
* |
I |
* |
|
|
|
| Emiliania huxleyivirus CAZ69528
Enterobacteria Phage ACT66763
Human parainfluenza virus 1 CAA26576 |
|
* |
* |
* |
* |
I |
|
|
|
|
Salmonella phage YP_003090232 |
|
|
|
* |
* |
I |
* |
* |
* |
|
| Lactobacillus Phage YP_002790822
Norovirus ADE28700(Numerous strains)
Norwalk-like virus Epiphyas postvittana
NPV
BAF38399 |
|
|
|
* |
* |
I |
* |
* |
|
|
Escherichia Phage YP_002003667
Bacillus phage S NP_690718
Mumps Virus AAD56373
GB virus A NP_045010 |
|
|
|
|
* |
I |
* |
* |
* |
|
| Staphylococcus phage phi YP_002332525
Enterobacteria phage TLS YP_001285537
Ectromelia virus NP_671600
Infectious spleen and kidney necrosis
virus NP_612246.
Vaccinia virus ABD52564
Variola Virus CAA53837
Variola Minor Virus CAB54683 |
|
|
|
|
|
I |
* |
* |
* |
* |
| Melanoma antigen Homo HSV-8 Gb Gb like
Homo sapiens |
|
|
Epi |
E |
E |
L |
|
|
|
|
| GAD2 Homo sapiens |
|
|
|
Epi |
E |
L |
E |
|
|
|
| Porcine circovirus
West Nile virus
Nicotinic receptor Homo |
|
|
|
|
Epi |
L |
E |
E |
|
|
| Bacillus Phage NP_690718
Mycobacterium Phage YP_002224980
Pseudomonas phage F116
YP_164316 Influenza B virus (Several strains) Rhodococcus phage ADD81097 |
|
* |
* |
* |
* |
L |
|
|
|
|
Influenza A ABO32793
|
|
|
|
* |
* |
L |
* |
* |
|
* |
Dugbe virus NP_690576
Feldmania species virus ACH46782
Hepatitis C virus subtype2a: AAF25612
Hepatitis C virus (isolate JFH-1)
BAB32872
Aeromonas Phage YP_656454 Staphylococcus phage 29NP YP_240601
Enterobacteria phage RB69
NP_861733 |
|
|
|
|
* |
L |
* |
* |
* |
|
Natrialba phage PhiCh1
NP_666010 |
|
|
|
* |
* |
L |
* |
* |
* |
|
Hepatitis C ACX44366
Enterococcus phage YP_001504324
Pseudomonas phage
YP_001956942
Streptomyces phage NP_813724 |
|
|
|
* |
* |
L |
* |
* |
|
|
Enterobacteria phage YP_277477
Lactobacillus phage YP_002117687
Lactococcus phage YP_762602
Staphylococcus phage SAP-2: YP_001491531
Human herpesvirus 4 type 2 YP_001129444
Human herpesvirus 5 ABV71532
Lymphocystis disease virus 1 Mammalian orthoreovirus
NP_694609
NP_078757Pseudomonas phage
YP_418089. Staphylococcus phage 85
YP_239732 Human papillomavirus
ACS72929 |
|
|
|
|
|
L |
* |
* |
* |
* |
Influenza A virus (Queensland ABO32793
Hepatitis C ACX44366
Enterococcus phage YP_001504324 |
|
|
|
* |
* |
L |
* |
* |
|
|
Amsacta moorei entomopoxvirus 'L'
Influenza Virus B (Russia ABQ81845) (Singapore ABL77016) and others (Connecticut, Memphis, Texas, Vienna, Romania)
Streptococcus
pyogenes phage NP_795674 |
|
|
* |
* |
* |
L |
* |
|
|
|
Table 5 The effects of the APP717 mutations on homology to bacterial
proteins. Protein accession numbers are provided and amino acid matches are
indicated by the asterisks or by the red letter of the mutant amino acid.
Commensal or common soil species are highlighted in bold.
Native 180 protein Hits |
I |
A |
T |
V |
I |
V |
I |
T |
L |
V |
Ricketsia canadensis A8EYN4 |
* |
* |
* |
* |
* |
V |
* |
* |
|
|
Aliivibrio salmonicida B6EPV1
Bacillus coagulans C1PC89
Rhodococcus Q0S6X2
Vibrio Metshnikovii C9P7P7 |
* |
* |
* |
* |
* |
V |
* |
|
|
|
Chlorobium ferrooxidans Q0YSY2
Hoeflea phototrophica A9CXL6
Rickettsia canadensis A8EYN4 (Plants)
Silicibacter pomeroyi Q5LMG7 |
|
* |
* |
* |
* |
V |
* |
* |
|
|
Ralstonia Eutropha Q472E0 (infects plants)
Ralstonia Picketii C6BIS4
Ralstonia solanasereum B5SKC7
Staphylococcus warneri C4W9N9 |
|
|
* |
* |
* |
V |
* |
* |
* |
* |
Acinetobacter baumannii A3M8V0
bacterium Ellin514 B9XI38 Carboxydibrachium pacificum B7RAI3 Clostridium asparagiforme C0CUU7
Erwinia tasmaniensis B2VL62
Lactobacillus Brevis Q03NB9
Lactobacillus crispatus D4FF71
Oxalobacter formigenes C3XCI5
Ralstonia solanacereum Q8XYV2:
Ryzobium
Salmonella enterica B5QA19
Salmonella heidelberg B5P6H1
Salmonella typhimurium A9N7S6 Thermoanaerobacter brockii C5UE75 Thermoanaerobacter congensis Q8R7X3 Thermoanaerobacter ethanolicus C7IUT0 Thermoanaerobacter pseudethanolicus B0KCL9
Thermoanaerobacter mathranii C6Q925
Vibrio fischeri B5EV74 |
|
|
* |
* |
* |
V |
* |
* |
* |
|
Carboxydibrachium pacificum
Clostridium asparagiforme C0CUU7 Clostridium bartlettii B0A6Z5
Clostridium thermocellum A3DFT3
Geobacillus sp. D3EBU3
Halothermothrix oreni B8CZE2
Maricaulis maris Q0AK61
Oligotropha carboxidovorans :B6J9Z4
Ralstonia pickettii B2UHX3
Sinorhizobium medicae A6U9B8
Tsukamurella paurometabola C2AKX9
Yersiniarohdei C4UTM6
Yersiniamollaretii C4SBZ5 |
|
|
|
* |
* |
V |
* |
* |
* |
* |
Mutant F 859 protein hits |
I |
A |
T |
V |
I |
F |
I |
T |
L |
V |
Aerococcus viridans D4YI11
Brachyspira hyodysenteriae C0QX46
Brevibacillus brevis C0ZA23
Clostridium butyric C4IJA0
Clostridium thermocellum D1NR57
Lactobacillus helveticus C9M4P6
Lactobacillus sakei Q38YE0
Oceanicola granulosus Q2CH54
Rhodobacterales bacterium A3VMF7
Vibrio fischerii B5FCH7 |
* |
* |
* |
* |
* |
F |
* |
|
|
|
Chloroherpetonia thalassium B3QTP3
Enterococcus faecalis Q838I9 (many strains)
Bacteroides vulgatus D4V0E1 |
|
* |
* |
* |
* |
F |
* |
* |
|
|
Vibrio Fischerii Q5E6H3 |
* |
* |
* |
* |
* |
F |
* |
|
|
|
Blautia Hansenii C9L542
Jannaschia sp Q28MI1
Providencia rettgeri D4BTW4
Providencia rustigianii D1NYV5
Rhodobacter sphaeroides Q3IZP4
Yersiniaintermedia C4T272 |
|
|
* |
* |
* |
F |
* |
* |
* |
|
Enterobacter cloacae D5CJ25
Idiomarina loihiensis Q5QU47
Proteus mirabilis C2LG18
Proteus penneri C0B3I7
Providencia alcalifaciens B6XHS9
Providencia stuartii B2Q1T7
Vibrio Furnissii C9PCZ1 |
|
|
|
* |
* |
F |
* |
* |
* |
* |
Mutant G 433 protein Hits |
I |
A |
T |
V |
I |
G |
I |
T |
L |
V |
Psychrobacter arcticus Q4FTU9 |
* |
* |
* |
* |
* |
G |
* |
* |
* |
|
Psychrobacter arcticus Q4FTU9
Streptosporangium roseum D2AZ15 |
|
* |
* |
* |
* |
G |
* |
* |
* |
|
Azorhizobium caulinodans A8HZ46
Bacillus mycoides C3AFH5
Catonella Morbia C4FVR1
Desulfuromonas acetoxidans Q1K1U7 Desulfovibrio Sp D2L952
Desulfovibrio magneticus C4XPD3
Dialister invisus C9LN97
Fusobacterium Varium C6JJE4
Granulibacter bethesdensis Q0BPF8
Hydrogenivirga sp A8V376
Legionella longbeachae D3HQH8
Leuconostoc citreum B1MYU7
Marinobacter aquaeolei A1U1N5 marine gamma proteobacterium A0Z7W7
Methylophaga thiooxidans C0N9Z9
Microcystis aeruginosa B0JM06
Nitrosomonas europaea Q82TG1
Photorhabdus asymbiotica C7BLV9
Vibrio alginolyticus D0X1F5
Vibrio harveyi A7MSV9
Yersinia pseudotuberculosis B7UF92 |
* |
* |
* |
* |
* |
G |
* |
|
|
|
Alcanivorax borkumensis Q0VS59
Bacillus cereus C2Y6K4 (Endemic soil)
Bacillus selenitireducens A8VSL6
Bacillus thuringiensis :C3IF91
Bacillus thuringiensis serovar israelensis Q3EPC6 Colwellia psychrerythraea Q482L7
Cyanothece sp. B1WRV0
Desulfococcus oleovorans Y1531
Dickeya dadantii D2BW75
Grimontia hollisae D0I6P8
Marine algicola DG893
Marinobacter aquaeolei A1U1P8
Natranaerobius thermophilus B2A0H4
Photobacterium angustum Q1ZQS6
Photobacterium profundum Q1Z1I8
Prevotella timonensis D1W1B7
Tolumonas auensis C4LCT3 |
|
* |
* |
* |
* |
G |
* |
* |
|
|
Bacillus licheniformis D2AZ15
Blastopirellula MarinaA3ZUC0
Brevibacillus Brevis C0Z7B6
Clostridium phytofermentans A9KJY1
Porphyromonas gingivalis Q7MVP9
Stenotrophomonas maltophilia B4SHS1
Verrucomicrobiae Bacterium B5JCZ1 |
|
|
* |
* |
* |
G |
* |
* |
* |
|
Acidovorax avenae D1STI2
Acidovorax ebreus B9MID3
Aeromonas hydrophila subsp. A0KHV9
Anaeromyxobacter sp. A7HEH4
Azorhizobium caulinodans A8IHP8
Azospirillum sp :D3P7H8
Bradyrhizobium sp A5EP47
Clostridium bolteae A8S3D7
Clostridium perfringens Q8XMX7
Dichelobacter nodosus A5EXF8
Gemellahaemolysans C5NVY9
Legionella longbeachae D3HN24
Leifsonia xyli Q6AED0
Methylibium petroleiphilum A2SD15
Methylobacterium extorquens C7C7Z2 Methylococcus capsulatus Q608X6
Mycoplasma penetrans Q8EUZ6
Myxococcus xanthus Q1DG48
Nakamurella multipartita C8X6U6
Pantoea ananatis :D4GKU3
Photobacterium profundum Q6LPG3
Polaromonas naphthalenivorans A1VT83
Ralstonia solanacearum A3RYP0
Rhodopseudomonas palustris Q6N682
leguminosarum bv. Trifolii C6AXC8
Rhizobium etli B3PP68
Rhizobium leguminosarum bv. Viciae Q1MBB9
Rhizobium meliloti Q92T03
Salinispora arenicola A8M7N9
Shewanella putrefaciens A2V1T8
Sinorhizobium medicae A6UEY9
Stenotrophomonas sp. B8KYQ9
Stenotrophomonas maltophilia B4SHS1
Syntrophobacter fumaroxidans A0LQ89
Thermosynechococcus elongatus Q8DHF1
Thermosipho melanesiensis A6LLD4
Thermotoga maritima Q9WYD7 |
|
|
|
* |
* |
G |
* |
* |
* |
* |
Mutant I 158 Protein Hits |
I |
A |
T |
V |
I |
I |
I |
T |
L |
V |
| Citrobacter
oseri A8AEH8
Citrobacter rodentium D2TQJ9
Clostridium phytofermentansA9KJW1
Desulfobacterium autotrophicum C0QE66
Enterobacter sp A4WC96
Escherichia coli Q8FG29 (many
strains)
Escherichia ferguson B7LUH8
Parachlamydia acanthamoebae D1R803
Shigella boydii Q323G5
Shigella dysenteriae B3WWP7
Shigella flexneri Q83R02
Veillonella dispar C4FS24
Yersinia intermedia C4T4Q3 |
* |
* |
* |
* |
* |
I |
* |
|
|
|
Bacillus cereus A7GLY6
Clostridium Humboltae A8RT62
Escherichia coli Q8X7P3
Grimontia hollisae D0I556
Haemophilus parasuis B8F464
Porphyromonas gingivalis B2RJP3
Staphylococcus aureus Q99RA3 Many strains
Xenorhabdus bovienii D3UZ08 |
* |
* |
* |
* |
* |
I |
|
|
|
|
Lactobacillus brevis Q03NB9
Lactobacillus iners C8PC46
Vibrio metschnikovii C9P8Z2 |
|
* |
* |
* |
* |
I |
* |
* |
* |
|
Clostridium perfringens Q8XLI1
Leptospira biflexa B0SNI9
Pedobacter heparinus C6Y2P1
Sphingobacterium spiritivorum C5PSB0 |
|
* |
* |
* |
* |
I |
* |
* |
|
|
Erwinia amylovora D4HU72
Erwinia pyrifoliae D2TBW3
Listeria innocua
Phytoplasma australiense B1V943 |
|
|
* |
* |
* |
I |
* |
* |
* |
|
Bacteroides sp D0TSW3
Bacteroides ovatus A7M5Q2_
Borrelia garinii B7XSM5
Clostridium difficile Q186M9
Clostridium cellulolyticum :B8I278
Corynebacterium urealyticum B1VG78
Pasteurella dagmatis C9PSK3
Proteus mirabilis C2LEY0_
Streptobacillus moniliformis D1AXI5 |
|
|
|
* |
* |
I |
* |
* |
* |
* |
Mutant L 198 Protein Hits |
I |
A |
T |
V |
I |
L |
I |
T |
L |
V |
Acidobactium Q1IJL1
Aggregatibacter actinomycetemcomitans D4ED73
Aggregati bacteria aphrophilus C6AKL6
Abiotrophia defectiva C4G6M0
Azorhizobium Caulinodans A8HQR6
Bacillus megaterium D5DSJ5
Bacillus pumilus B4ADS6
Bdellovibrio bacteriovorus Q6MRD1
Caulobacter crescentus B8H097 Clostridiales bacterium C5EIM1
Clostridium butyricum C4ICE7
Clostridium methylpentosum C0ECW3
Chlamydophila abortus Q5L6C5
Coprococcus Comes C0BFC3
Doreaformici generans B0G8H5
Eikenella Corrodens C0DY03
Mycobacterium sp Q1BC56
Roseobacterium denitrificans Q162L0
Ruminococcus obeum D4LS87 |
* |
* |
* |
* |
* |
L |
* |
|
|
|
Carnobacterium sp A8UAH5
Mycobacterium abscessus B1MP36
Mycobacterium chelonae A5A9P9 |
|
|
* |
* |
* |
L |
* |
* |
* |
* |
Bacilluscereus lus Rock3-44 C3AYE2
Bacillus thuringiensis Q3EUN8_ |
|
* |
* |
* |
* |
L |
* |
* |
* |
|
Bacillus mycoides C3AYE2
Bacillus pseudomycoides C3BEV6
Gramella forsetii A0LYP6
Lyngbya sp A0YPN4
Lysinibacillus sphaericus B1HWG7 |
|
|
* |
* |
* |
L |
* |
* |
* |
|
Butyrate-producing bacterium D4MSZ1
Citreicella Sp. D0D4G6
Clostridium sp. D4CDQ6
Escherichia coli B5AXC7
Leifsonia xyli subsp.xyli Q6AFD7 Mycoplasma Pulmonis MYPU_0850 Neptuniibacter caesariensis :Q2BPQ0 Rhodopseudomonas palustris Q21C19_
Ruminococcus torques A5KJD0
Staphylococcus hominis C2LWL2
Veillonella dispar C4FNF7
Veillonella parvula D1BQP4
Yersiniarohdei C4UUS1
Yersinia frederiksenii C4SLT3 |
|
|
|
* |
* |
L |
* |
* |
* |
* |
Table
6 The effects of the Swedish
APP mutation on homology to viral proteins. Protein accession numbers are
provided and amino acid matches are indicated by the asterisks or by the
red letter of the mutant amino acid. Species with proteins containing epitopes
matching those of the peptde amino acids are also shown (Marked by E) Phages
infecting commensal bacteria and common viruses (eg Rhinoviruses and influenza)
are highlighted in bold.
Virus |
T |
E |
E |
I |
S |
E |
V |
K |
M |
| Simian
immunodeficiency virus 239
APP Homo sapiens |
|
|
|
|
|
Epi |
E |
E |
E |
| Influenza
A virus Chicken
BAH03519 Goose
ABJ96738 Duck
ABJ96735 |
|
* |
* |
* |
* |
* |
* |
* |
|
| Synechococcus phage S-PM2 YP_195120 |
|
* |
* |
* |
* |
* |
* |
|
|
| Influenza Rio ADE75392 |
|
* |
* |
* |
|
* |
* |
* |
|
Influenza A virus Hong Kong BAJ07997
Influenza A virus Egypt ADG21445 and many Avine influenza viruses |
|
* |
* |
* |
* |
|
* |
* |
|
Bacillus phage NP_046598 |
|
* |
* |
* |
* |
* |
|
|
* |
Bornavirus ADB84600 |
|
|
* |
* |
|
* |
* |
|
* |
Spiroplasma phage SVGII3 CAI94596 |
|
|
|
|
* |
* |
* |
* |
* |
Synechococcus phage Syn5 ABP87933 |
|
|
|
|
|
|
* |
* |
* |
Listeria phage YP_002261477
Aeromonas phage NP_944230
Enterobacteria phage AAN28262 |
|
|
|
|
|
* |
* |
* |
* |
Influenza B virus ABL76812 (Both) |
|
* |
* |
* |
* |
|
|
|
|
Influenza B virus ACF54229 (Both) |
|
|
|
* |
* |
* |
* |
|
|
| Chlamydia trachomatis
Homo sapiens protein tyrosine phosphatase, receptor
Human papillomavirus type 16
Human papillomavirus type 6b
Human respiratory syncytial virus |
|
|
|
|
|
Epi |
E |
N |
L |
Influenza Rio ADE75392 |
|
* |
* |
* |
- |
* |
* |
- |
L |
Mycobacterium phage ADA83789
Streptococcus phage NP_056710 |
|
|
* |
* |
* |
* |
- |
N |
L |
| Bacillus phage TP21-L YP_002333578
Borna virus ACG59353
Campylobacter phageCBJ94264
Corynebacterium phage YP_001468961
Enterobacteria phage Fels-2 YP_001718743
Enterobacteria phage phiEco32 YP_001671828
Liao ning virus YP_460028
Menangle virus YP_415513
Synechococcus phage S-PM2 YP_195115
Unidentified Fusellovirus AAL24816 |
|
|
|
* |
* |
* |
* |
N |
|
Synechococcus phage YP_195115 |
|
|
* |
* |
* |
* |
* |
N |
|
| Respiratory syncytial virus ACO83301
Human adenovirus 13 BAG48790,
17 ABQ00199 , 38 BAG48815,
39 BAG48816, 47 BAG48824
Hantavirus Yakeshi-Mm-59 AAB22506
Pseudomonas phage LUZ24
YP_001671894
Puumala virus ABO09803
Muju virus ABM92784
Dobrava-Belgrade virus ADA68892
Flanders virus AAN73285
Clostridium botulinum D phage ZP_04863764
Tacaribe virus NP_694849 |
|
|
|
|
* |
* |
* |
N |
L |
| Borna Virus
ACG59353 |
|
|
|
|
* |
* |
* |
N |
|
| Acanthamoeba
polyphaga mimivirus YP_001426695
Human adenovirus 41 AAA42440
Beilong virus AAA42439
Fer-de-lance virus NP_899661
Pichinde virus ABU39911
Influenza
virus Illinois ABI83868|
Rhinovirus C ACR14890
Human rhinovirus 28, ABF51202
Human Rhinovirus 68, ACU27211
and Rhinovirus .sp |
|
|
|
|
|
* |
* |
N |
L |
Enterobacteria phage AR1 BAI83009
Synechococcus phage YP_003097343
Enterobacteriaphage YP_002853956
Streptococcus phage 858 YP_001686831
Enterobacteria phage T4 BAA23634
Enterobacteria phage AR1 BAI83009
Synechococcus phage S-RYP_001957167 201phi2-1p448
Pseudomonas phage YP_001957167
Enterobacteria phage RB32 YP_238518
Streptococcus phage 2972 YP_238518
Bordetella
phage NP_958690 |
|
|
|
|
|
* |
* |
N |
L |
|
Table 7 The effects of the Swedish
mutations on homology to bacterial proteins. Protein accession numbers are
provided and amino acid matches are indicated by the asterisks or by the
red letter of the mutant amino acid. Commensal or common soil bacteria are
highlighted in bold.
Bacteria |
T |
E |
E |
I |
S |
E |
V |
K |
M |
| Atopobium
Parvilum C8WAI5 |
* |
* |
* |
* |
* |
* |
* |
* |
|
| Bermanella
maris Q1N288
Shewanella violacea
D4ZM52 |
|
* |
* |
* |
|
* |
* |
* |
* |
| Vibrio
furnissii C9PJ26
|
|
|
* |
* |
* |
* |
* |
* |
* |
Lactis subsp Q031F7
Lactocococcus cremoris
A2RIQ5 Same
for both |
* |
* |
* |
* |
* |
* |
|
|
|
Bacillus anthracis C3PD86
Bacillus thuringiensis C3HJR4
Bacillus thuringiensis serovar pondicheriensis C3GK36
Baciillus Cereus C2THS7 several strains |
|
* |
* |
* |
|
* |
* |
* |
* |
| Clostridum
tetani Q896K9
Oceanobacillus Q8CV16 |
|
* |
* |
* |
* |
* |
* |
* |
|
Colwellia psychrerythraea Q487K7
Enterococcus Faecium D3LK32
Ricketsia Typhi Q68XP3
Rickettsia Prowazekii O05942
Photorabdus luminens Q7N0P2 Same for both |
* |
* |
* |
* |
* |
* |
* |
|
|
| Streptobacillus
moniliformis
D1AV81 |
|
* |
* |
* |
* |
* |
* |
* |
|
Leptotrichia Buccalis
C7NE35
Rumibococcus Sp C6JEQ4
Streptococcus pneumoniae
ABJ55426
Sams for both |
|
|
* |
* |
* |
* |
* |
|
|
Clostridium beijerinckii A6LY25 Sanme for both
|
* |
* |
* |
* |
* |
* |
* |
|
|
Planctomyces maris A6C251
|
|
|
|
* |
* |
* |
* |
* |
* |
| Leptospira
biflexa B0SKY9 Sane for both
|
|
* |
* |
* |
* |
* |
|
|
|
| Mycoplasma
hominisD1J7S2
|
|
|
* |
* |
* |
* |
* |
* |
|
Sodalis glossinidius Q4LC19 Same for both
|
* |
* |
* |
* |
* |
* |
|
|
|
| Nodularia
spumigena A0ZE12 Same for both
|
* |
* |
* |
* |
* |
|
|
|
|
| Streptococcus
pneumoniae ZP_01821444 Same for both |
|
* |
* |
* |
* |
* |
|
|
|
|
T |
E |
E |
I |
S |
E |
V |
N |
L |
Fusobacterium sp C3WJ64
|
|
* |
* |
* |
|
* |
* |
N |
L |
Fusobacterium mortiferum C3WDH2
|
|
* |
* |
* |
* |
|
* |
N |
L |
Acidobacterium psulatum C1F5P2
|
* |
* |
* |
* |
* |
* |
|
N |
L |
Corynebacterium matruchotii C5VDU4
Escherichia coli O1 57 H7 Q8X597
Escherichia coli O6 Q8FGX7
Escherichia coli (strain UTI89 / UPEC) Q1RB23
Escherichia coli O6 Q0TH59 and numerous other E.Coli
strains
Shigella flexneriQ83RH3 Helicobacter
hepaticus Q7VIZ4
Staphylococcus haemolyticus Q4L6V6
Trichodesmium erythraeum Q10V04 |
|
* |
* |
* |
* |
* |
* |
N |
|
Bacteroides thetaiota Q8A9X8
Anaerococcus lactolyticus C2BI02
|
|
|
* |
* |
* |
* |
* |
N |
L |
Synechococcus sp. Q7U980
|
|
* |
* |
* |
* |
* |
* |
- |
L |
Listeria grayi C2BZ78
Streptococcus pneumoniae ZP_01817518 |
|
|
|
* |
* |
* |
* |
N |
L |
Citrobacter Youngae D4B9G4
Pseudoalteromonas atlantica Q15N87
Pseudomonas
chlororaphis P31521
Pseudomonas fluorescens C3K287
Rhodobacter sphaeroides A4WVX1
Robiginitalea biformata A4CHQ4
Streptococcus pneumoniae CAI34213 |
|
|
|
|
* |
* |
* |
N |
L |
Streptococcus pneumoniae ZP_02717209 |
|
|
|
* |
* |
* |
* |
N |
|
Table 8
Viral proteins lining up with autoantigens from Chronic obstructive pulmonary
disease, myasthenia gravis, multiple sclerosis and pemphigus vulgaris and
to mutant proteins from polyglutamine repeat disorders (Huntington’s disease,
Dentatorubropallidoluysian atrophy, Kennedy disease and Spinocerebellar ataxias)
and from cystic fibrosis. APOE4 is included as an example of a risk factor
in a number of polygenic diseases. Accession numbers and the aligning sequences
are shown with references where the virus has been implicated in the relevant
disease. The polyglutamine expansions also increase the antigenicity of the
resultant peptide with each triplet QQQ addition.
| Autoantigen |
Viral Homolog |
Sequence |
Involvement |
| Myaesthenia
Gravis Nicotinic receptor CHRNG NP005190 acetylcholine receptor subunit
gamma
|
ABC71426 Human echovirus 18
AF427971 Human coxsackievirus B1 |
FDWQNC
|
? |
| AF507093Bovine rotavirus G10
|
IVVNA |
? |
| CAF24822 Human herpesvirus 1
ABW83347 Human herpesvirus 2 |
R-R--RDY-G-
VLRV
|
Anti nicotinic receptor antibodies
isolated from myasthenia patients cross react with HSV-1 glycoprotein
D (119) |
| ACC94304 Tamiami virus |
EEALTT |
? |
| NP_477882 Shrimp white spot
syndrome virus |
DIVLEN |
? |
| ADD94092 uncultured phage |
NQEERL |
? |
| Ssystemic
lupus erythematosus XRCC6 NP_001460 |
ADD65207.1 glycoprotein
Sandfly fever Naples virus] AAN06959.1
polyprotein Toscana virus] |
TLFSALLI
|
? |
| >emb|CAA37487.1|
hypothetical protein [Non-A, non-B hepatitis virus] |
VPQEEEL
|
? |
| BAA77426.1 ORF2
Torque teno virus] |
LLR-VRAK
|
? |
| ACS29433.1 polyprotein
Hepatitis C virus] |
RDSLI-LV
|
Lupus is frequently
reported in Hepatitis C patients {Ramos-Casals, Munoz, et al. 2009 1801
/id} |
| AAA52748.1 polyprotein
Hepatitis C virus subtype 1b] |
PPDYNP
|
| NP_149906.1 443R Invertebrate
iridescent virus 6] |
TRTF-TSTG
And
DIISIA---DL |
? |
| AAC35885.1 protease
bovine adenovirus 10] |
SEEELK-HI
|
? |
| Pemphigus
Vulgaris Desmoglein-3 NP_001935
|
Human herpesvirus 1 |
VNKTIT |
Occasionally associated with
HSV-1 (120) |
| ABB22271 Ovine herpesvirus 2
|
STGGTN |
? |
| AAP82014 Suid herpesvirus 1
|
TGALAI |
? |
| CBJ94251 Campylobacter phage
|
KFKKLA |
? |
| AAX8491 Xanthomonas phage |
K-LVDYIL |
? |
| YP_001950153 Ralstonia phage
|
QLRGSHT |
? |
| NP_958691 Bordetella phage |
SGQSGTM
|
? |
| NP_112040 Enterobacteria phage |
IEGAHPE
|
? |
| Chronic
obstructive pulmonary disease Elastin NP_000492.2 elastin isoform a
precursor
|
ABI15777. Encephalomyocarditis
virus
NP_653077 Equine rhinitis B virus
ABB97066 Mengo virus
|
GLPYTT
|
? |
| ACI22601 Influenza
B virus |
SILH-SRP
|
Picornavirus ,influenza;
respiratory syncytial virus; corona viruses, parainfluenza; adenovirus
and human metapneumovirus detected during COPD exacerbation (121) |
| BAE96916 Respiratory syncytial
virus |
Y-AAKA
A
|
| AAA47928 Theiler's encephalomyelitis
virus |
PGFGL-P
|
? |
| NP_044050 Molluscum contagiosum
virus subtype1 |
IFPGGA
|
? |
| BAH15164 Serratia phage |
GLSPIF
|
? |
| CAM12729 Zucchini yellow mosaicvirus
|
VGGIPT
|
? |
| ADF28539 Human TMEV-like cardiovirus
:
ACO92355 Saffold virus |
PGFGLS
|
? |
| ABD73306 Gremmeniella abietina
type B RNA virus |
PIKAPKL
|
? |
| NP_612874 Clostridium
phage |
G+GLPYTT
|
? |
| BAA03030 Orgyia pseudotsugata
single capsid nuclopolyhedrovirus |
APRPGV |
? |
| Multiple
sclerosis Crystallin CRYAB
|
EBNA3 Epstein-Barr virus |
DQFFG |
Implicated
(122) |
| D2XR26 Bacillus
phage |
HSPSRL |
? |
| 9PARAQ84747 Human
parainfluenza virus 1 |
R-PSFLR |
Implicated
(123) |
| C9WSX19 Norovirus dog |
SLSSDGV |
Dog ownership has been
linked to multiple sclerosis (124;125) |
| Multiple
sclerosis Myelin associated glycoprotein MAG
AAB58805
|
YP_164320 Pseudomonas phage
|
AEDGVYA
|
? |
| YP_003406894 Marseille
virus |
KYYFRG
|
? |
| CAA24862.1 Human herpesvirus
4 (Epstein Barr) |
PAVLGR--E
|
Implicated
(122)
|
| P25939 Human herpesvirus 4 (Epstein
Barr) |
LRGQ-QAP
|
| CAA66845 Human herpes virus
4 (Epstein Barr) |
DEGTWV
|
| AAD51697 Human herpesvirus 4
(Epstein Barr) |
PSSIS
|
| ABV71654.Human herpesvirus 5
(Cytomegalovirus) |
CP+LRP-LS-
L |
Implicated: Seropositivity
predicts a better outcomein Multiple Sclerosis (Beneficial virus ?)
{Zivadinov, Nasuelli, et al. 2006 1800 /id} |
| AAR31274 Human herpesvirus 5
(Cytomegalovirus) |
GKYYF-R-D |
| YP_015543 Pyrobaculum spherical
virus |
YITQTR
|
? |
| Multiple
Sclerosis MBP NP_001020252 myelin basic protein isoform 1
|
ACO87999 Hirame rhabdovirus |
GRGLSL |
? |
| ADE45455 Polyomavirus
HPyV7 |
RSGSPM |
? |
| YP_073799 Human herpesvirus
7 |
PSQRHGS |
Detected in MS tissue (126)
|
| NP_042301 Southern cowpea mosaic
virus |
PGRSPLP
|
? |
| YP_143172 Acanthamoeba polyphaga
mimivirus |
IGRFFGG
|
? |
| ABB22292 Ovine Herpesvirus
2 NP_065571 Alcelaphineherpes virus1 |
FFKNIV
|
? |
| JC polyomavirus |
PRTPPP
|
Seropositivity in some Multiple
sclerosis patients (127) |
| BAA00490 Ornithogalum mosaicvirus |
TQDENP
|
? |
| YP_003280846 Helicobasidium
mompa endornavirus1 |
DSIGRF
|
? |
| CAA32420 Simian rotavirus |
ARTAHY
|
? |
| YP_002117760
Pseudomonas phage |
IVTPRT
|
? |
| YP_164431 Bacillus phage |
GRASDY
|
? |
| NP_570206 Swinepoxvirus ACV04605
Nakiwogovirus |
TLSKIF
|
? |
| YP_214645 Prochlorococcus phage |
GRSPLP
|
? |
| YP_142835 Acanthamoeba polyphaga
mimivirus |
LDSIGR
|
? |
| ACZ8140 Moussavirus |
TAHYGS
|
? |
| BAA77241 Broadbean wilt virus2 |
RASDYK
|
? |
| Multiple
sclerosis Myelin Oligodendrocyte protein MOG
|
NP_944019 Aeromonasphage |
AMELK |
? |
| ADA81168 Staphylococcus phage |
VLGPLV |
? |
| NP_690686 Bacillus phage |
LVALII |
? |
| ABO87130 Hepatitis delta virus
|
KDQDG |
? |
| ADE60693 Rice stripe virus |
SRVVHL
|
? |
| ACA24946 Swine parainfluenzavirus3
|
RDHSY
|
? |
| AAL89267 Shrimp whitespot syndrome
virus |
NLHRTF
|
? |
| NP_569759 Mycobacterium phage
|
ELLKDA+G |
? |
| Multiple
sclerosis NP_000524
myelin
proteolipid protein isoform 1
|
BAB83467 Chlorellavirus |
SATVTGGQ |
? |
| CAG70345 Bovine viral diarrhea
virus 1 |
VPVYIY
And
GITYA
|
? |
| YP_001111042Burkholderia phage
|
FNTWT |
? |
| AF310938 Powassanvirus
NP_620108 Langatvirus |
CSAVPV |
? |
| AAW33310 Human adenovirus4 |
GLLEC |
Associated with relapse in multiple
sclerosis (128) |
| ABO42303 Avian meta pneumo virus
|
KTSA-SIGSLC
|
? |
| ABU82778 Human metapneumovirus
|
KTSA-SIGSLC
|
? |
| YP_002241563 Mycobacterium
phage Butterscotch
YP_002241480 Mycobacterium phage Troll4
YP_655259 Mycobacterium phage PBI1 |
KNYQDY
|
? |
| YP_717772 Synechococcus phage |
YQDYEY |
? |
| YP_001129421 Human herpes virus
8 (Kaposi’s sarcoma virus) |
PFASL-A
And NFAVL |
? |
| ABI35813 Human meta pneumovirus
|
KTSA-SIGSLC |
? |
| YP_002241961 Mycobacterium phage
Gumball
YP_001936156 Mycobacterium phage Adjutor |
KNYQDY
|
? |
| AAK69175 Bovine viral diarrhea
virus 1 |
VPVYIY
|
? |
| YP_71777 Synechococcus phage
syn9 |
YQDYEY
|
? |
| ABB89216 Human herpes virus
4 (Epstein Barr) |
AHSLE
|
Implicated
(122) |
| Polyglutamine
repeats, Huntinhton’s disease, spinocerabellar ataxias, Dentatorubropallidoluysian
atrophy, Kennedy disease Querey = qqqqqqqqqqqqqqqqqqqqqqq |
AAS86764 Zantedeschia mild mosaic
virus |
QQQQQQQQQQQQQ-QQQQQQQQQ
|
| AAQ96572 Vibrio phage VP16C
|
QQQQQ--------QQQQQQ----QQQQQQQQQQQQ
|
| YP_001468520
Listeria phage A511 |
QQQQ----------QQQ---QQQ--QQQ--QQ--------QQQQQQQ
|
| YP_398993 Enterobacteria Phage |
QQ---QQQQQQQQQQQQQQQQQQQ
|
| gbAAC57158 Human herpesvirus
8 type M |
+QQQQ+QQQQ—QQQQ—QQQQ--QQQQ |
| AAR14310
Lactococcus phage ul363 |
Q-QQQQQQ----Q-Q---Q-Q-QQ---QQQ+Q
|
| YP_001456769
Corynebacterium phage |
QQQ--Q+Q----QQQ--Q+Q-----QQQQ-----QQQ
|
| NP_116331 T2 Tupaiid herpesvirus
1 |
QQQQQQQQQQQQQQQQQQQQQQQ
|
| ACO25273 Epizootic haematopoietic
necrosis virus |
+QQ++QQQQQQQQQQQQ++Q++Q |
| AAL89066 Shrimp white spot syndrome
virus |
QQQQQQQ-+QQQQ-+QQQQQQQQ
|
| YP_001218813
Pseudomonas phage |
QQ--Q-Q--QQ--QQQQQQQQ-Q
|
| embCAA52472
Human papillomavirus type 3 |
+QQQQQQQQQQQQQ+Q |
| CAA75466Human papillomavirus
type 77 |
QQQQQQQQQQQQQ
|
| Human adenovirus 1 |
++QQQQQQQQ++------Q+Q |
| AAA79432 Human papillomavirus
type 29 |
QQQQQQQ+QQQQQQQ
|
| CAA75466 Human papillomavirus
type 77 |
QQQQQQQQQQQQQ
|
| AAL50729 Human herpesvirus 6B
|
QQQQQ+QQQQ |
| NP_050183 Human herpesvirus
6 |
QQQQQ+Q-QQQ |
| ADB84736 Human herpesvirus 5
|
QQQQQQQ+Q
|
| ABQ51392 Human rhinovirus sp.
|
QQQ |
| Cystic
fibrosis CFTR
|
AAB59808 Vaccinia virus
CAA53834 Variola virus |
GT-IKENII---G |
|
| YP_003090182 Burkholderia phage
KS9 |
IIGVSY-DE |
Burkholderia infection has been
related to Cystic fibrosis (126;129) |
| YP_001504144 Enterococcus phage
|
DEYR-RSVI
|
|
| ACF59816 Human parainfluenza
virus 4b |
GTIKE-II
|
|
| YP_908809 Staphylococcus phage
YP_002003602 Escherichia phage rv5 |
KENIIG
|
|
| NP_046572 Bacillus phage
YP_052931 Palyam virus
BAA34933 Chuzan virus |
GTIKEN
|
|
| ABD63811Lactococcus
phage YP_717768 Synechococcus phage |
VSYDEY
|
|
| AAM4760 Dendrolimus
punctatus cypovirus 1 |
MPGTIK
|
|
| YP_00183702 Enterobacteria
phage |
TIKENI
|
|
| YP_003358383 Pseudomonas phage
YP_002456113 Erwinia phage |
GVSYDE
|
|
| Polygenic
diseases APOE4 gi15826311 pdb 1B68 A Chain A, Apolipoprotein E4 |
ACE82482 Hepatitis C virus subtype
1a |
GADMEDV
|
|
| ACT53109 Hantavirus Jurong ACO37156
Seoul virus |
ET+KELKA
|
|
| NP_932306 Botrytis virus X |
ALM-ETMK
|
|
| NP_116510 Lactococcus
phage |
K-EL+EQLTP
|
|
| AAN03856 Adeno-associated virus-8 |
LS-IRE-LGP
|
|
| YP_00292273 Burkholderia phage |
Q--WQSGQ |
|
| YP_002455799 Lactobacillus phage
|
MKELKA |
|
| YP_003358506 Shigella phage |
VQTL-EQV+E
|
|
| DAA06495 Human herpesvirus 5 |
DDL—R-LAVYQA
|
|
| ABI63513 Human herpesvirus 1 |
RLA-HLR
And KRLLR
|
Binds to APOE (see text) |
| AAK50002 Human herpesvirus 8 |
LA-S-LR-LRKR |
|
| AAT07716 Human herpesvirus 3
(Varicella Zoster) |
LEEQLT--A
|
|
| ACM48047 Human herpesvirus 5 |
LRD-D--DLQKR
|
|
| YP_001129382 Human herpesvirus
8 |
G+RWEL
And LVEQ--VR
|
|
| AAY41102 Human herpesvirus 4 |
G-D-EDVR
|
|
Figure 1
Figure 2
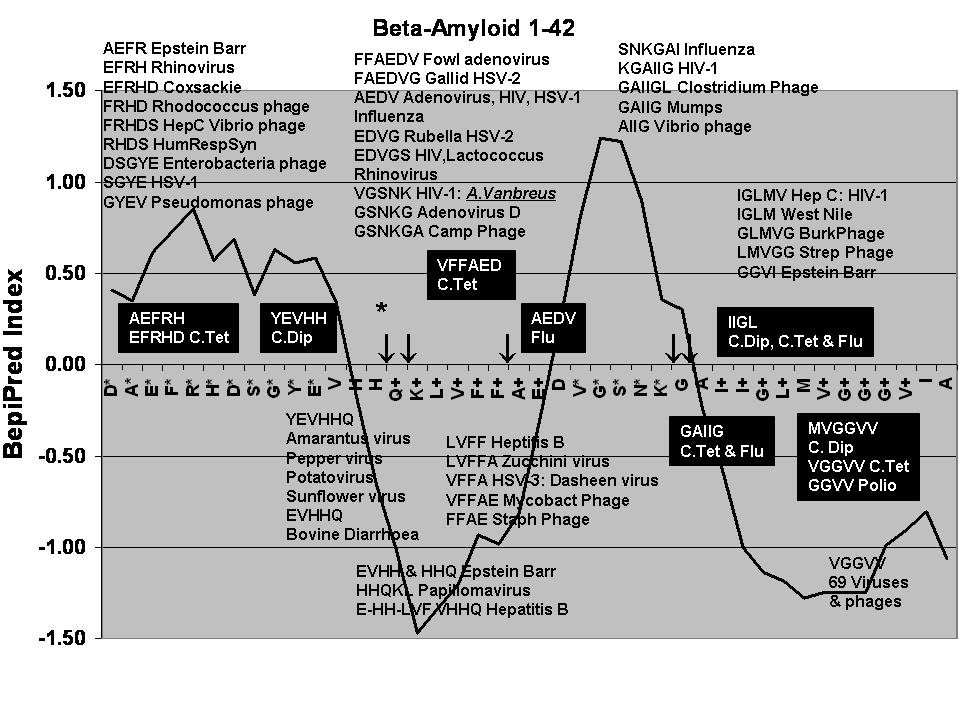
Viral protein matches to different regions of the beta-amyloid peptide in
relation to the predicted B Cell Epitope antigenicity. Predicted epitopes
are marked with an asterisk and the Y-axis is an index of antigenicity. The
VGGVV epitope has been used to label beta-amyloid and is marked + as are other
short epitopes used to label beta-amyloid (QKLV, FFAE, IIGL) Other short epitopes
used to label beta-amyloid include MGGVV, VGGVV, MVGGVV, VGGVV and GGVVIA).
The viruses and bacteria in black boxes represent those where vaccination
was reported to reduce the incidence of Alzheimer’s disease. These alignments
are to viral proteins rather than to epitopes within the vaccine. The arrows
represent the beta-amyloid cleavage sites of the catalytic beta-amyloid autoantibodies
isolated from Alzheimer’s disease sera. H*↑Q is the major site of catalysis.
Note that cancer and plant viruses overlap this region. Burk= Burkholderia
Phage; Camp
Phage = Campylobacer
Phage; HumRespSyn = Human respiratory syncitial Virus; HepC = Hepatitis C;
HIV = Human Immunodeficiency Virus; HSV-1 = Herpes simplex (Human Herpesvirus
1); Mycobact = Mycobacteria phage; Strep = Streptococcus phage; C.Tet = Clostridium
Tetani, C.Dip = Corynebacterium diphtheriae (See Table 2 for Accession numbers)
Figure 3
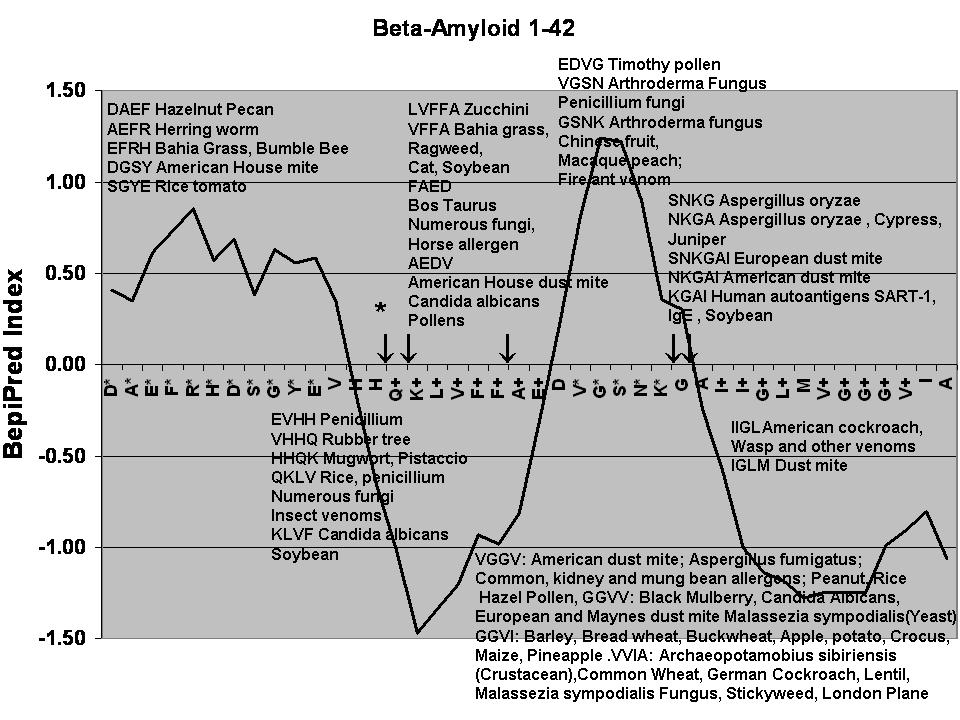
Allergenic protein matches to different regions of the beta-amyloid peptide
in relation to the predicted B Cell Epitope antigenicity. Predicted epitopes
are marked with an asterisk and the Y-axis is an index of antigenicity .Allergens
expressing proteins that match different regions of the beta-amyloid peptide
are aligned with their respective matches. The arrows represent the beta-amyloid
cleavage sites of the catalytic beta-amyloid autoantibodies isolated from
Alzheimer’s disease sera. H*↑Q is the major site of catalysis (see Table
2 for Accession numbers).
Reference List
(1) Kellner A, Matschke J, Bernreuther C, Moch
H, Ferrer I, Glatzel M. Autoantibodies against beta-amyloid are common in
Alzheimer's disease and help control plaque burden. Ann Neurol 2009 Jan;65(1):24-31.
(2) Paul S, Planque S, Nishiyama Y. Immunological
Origin and Functional Properties of Catalytic Autoantibodies to Amyloid beta
Peptide. J Clin Immunol 2010 May 8.
(3) Sohn JH, So JO, Hong HJ, Kim JW, Na DR,
Kim M, et al. Identification of autoantibody against beta-amyloid peptide
in the serum of elderly. Front Biosci 2009 Jan 1;14:3879-83.
(4) Itagaki S, Akiyama H, Saito H, McGeer PL.
Ultrastructural localization of complement membrane attack complex (MAC)-like
immunoreactivity in brains of patients with Alzheimer's disease. Brain Res
1994 May 9;645(1-2):78-84.
(5) McGeer PL,
Akiyama H, Itagaki S, McGeer EG. Activation of the classical complement pathway
in brain tissue of Alzheimer patients. Neurosci Lett 1989 Dec 15;107(1-3):341-6.
(6) Pyles RB. The association of herpes simplex
virus and Alzheimer's disease: a potential synthesis of genetic and environmental
factors. Herpes 2001 Nov;8(3):64-8.
(7) Schwab C, Akiyama H, McGeer EG, McGeer
PL. Extracellular neurofibrillary tangles are immunopositive for the 40 carboxy-terminal
sequence of beta-amyloid protein. J Neuropathol Exp Neurol 1998 Dec;57(12):1131-7.
(8) Morelli MA, DeBiasi M, DeStradis
A, Tamburro AM. An aggregating elastin-like
pentapeptide. J Biomol Struct Dyn 1993 Aug;11(1):181-90.
(9) Cribbs DH, Azizeh BY, Cotman CW, LaFerla
FM. Fibril formation and neurotoxicity by a herpes simplex virus glycoprotein
B fragment with homology to the Alzheimer's A beta peptide. Biochemistry 2000
May 23;39(20):5988-94.
(10) Itzhaki RF, Wozniak MA. Herpes simplex virus
type 1, apolipoprotein E, and cholesterol: a dangerous liaison in Alzheimer's
disease and other disorders. Prog Lipid Res 2006 Jan;45(1):73-90.
(11) Wozniak MA, Itzhaki RF, Shipley SJ, Dobson
CB. Herpes simplex virus infection causes cellular beta-amyloid accumulation
and secretase upregulation. Neurosci Lett 2007 Dec 18;429(2-3):95-100.
(12) Wozniak MA, Frost AL, Itzhaki RF. Alzheimer's
disease-specific tau phosphorylation is induced by herpes simplex virus type
1. J Alzheimers Dis 2009 Feb;16(2):341-50.
(13) Armien AG, Hu S, Little MR, Robinson N,
Lokensgard JR, Low WC, et al. Chronic Cortical and Subcortical Pathology with
Associated Neurological Deficits Ensuing Experimental Herpes Encephalitis.
Brain Pathol 2009 Nov 5.
(14) Letenneur L, Peres K, Fleury H, Garrigue
I, Barberger-Gateau P, Helmer C, et al. Seropositivity to herpes simplex virus
antibodies and risk of Alzheimer's disease: a population-based cohort study.
PLoS One 2008;3(11):e3637.
(15) Friedland RP, Tedesco JM, Wilson AC, Atwood
CS, Smith MA, Perry G, et al. Antibodies to potato virus Y bind the amyloid
beta peptide: immunohistochemical and NMR studies. J Biol Chem 2008 Aug 15;283(33):22550-6.
(16) Altschul SF, Madden TL, Schaffer AA, Zhang
J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation
of protein database search programs. Nucleic Acids Res 1997 Sep 1;25(17):3389-402.
(17) Ivanciuc O, Schein CH, Braun W. SDAP: database
and computational tools for allergenic proteins. Nucleic Acids Res 2003 Jan
1;31(1):359-62.
(18) Goodman R, Ebisawa M, Sampsom H, Van Ree
R, Vieths S, Wise J, et al. AllergenOnline, a peer-reviewed protein sequence
database for assessing the potential allergenicity of genetically modified
organisms and novel food proteins. World Allergy Organization Journal 2007;S285:285.
(19) Salimi N, Fleri W, Peters B, Sette A. Design
and utilization of epitope-based databases and predictive tools. Immunogenetics
2010 Apr;62(4):185-96.
(20) Larsen JE, Lund O, Nielsen M. Improved method
for predicting linear B-cell epitopes. Immunome Res 2006 Apr 24;2:2.
(21) Zhang T, Breitbart M, Lee WH, Run JQ, Wei
CL, Soh SW, et al. RNA viral community in human feces: prevalence of plant
pathogenic viruses. PLoS Biol 2006 Jan;4(1):e3.
(22) Colson P, Richet H, Desnues C, Balique F,
Moal V, Grob JJ, et al. Pepper mild mottle virus, a plant virus associated
with specific immune responses, Fever, abdominal pains, and pruritus in humans.
PLoS One 2010 Apr 6;5(4):e10041.
(23) Contini C, Seraceni S, Cultrera R, Castellazzi
M, Granieri E, Fainardi E. Chlamydophila pneumoniae Infection and Its Role
in Neurological Disorders. Interdiscip Perspect Infect Dis 2010;2010:273573.
(24) Miklossy J, Khalili K, Gern L, Ericson RL,
Darekar P, Bolle L, et al. Borrelia burgdorferi persists in the brain in chronic
lyme neuroborreliosis and may be associated with Alzheimer disease. J Alzheimers Dis 2004 Dec;6(6):639-49.
(25) Kountouras J, Boziki M, Gavalas
E, Zavos C, Grigoriadis N, Deretzi G, et al. Eradication
of Helicobacter pylori may be beneficial in the management of Alzheimer's
disease. J Neurol 2009 May;256(5):758-67.
(26) Kim JM, Stewart R, Prince M, Kim SW, Yang
SJ, Shin IS, et al. Dental health, nutritional status and recent-onset dementia
in a Korean community population. Int J Geriatr Psychiatry 2007 Sep;22(9):850-5.
(27) Stein PS, Desrosiers M, Donegan SJ, Yepes
JF, Kryscio RJ. Tooth loss, dementia and neuropathology in the Nun study.
J Am Dent Assoc 2007 Oct;138(10):1314-22.
(28) Hoffmann M, Muniz J, Carroll E, De Villasante
J. Cryptococcal meningitis misdiagnosed as Alzheimer's disease: complete neurological
and cognitive recovery with treatment. J Alzheimers Dis 2009 Mar;16(3):517-20.
(29) Ala TA, Doss RC, Sullivan CJ. Reversible dementia: a case of cryptococcal meningitis
masquerading as Alzheimer's disease. J Alzheimers Dis 2004 Oct;6(5):503-8.
(30) Veerhuis R, Janssen I, De Groot CJ, Van
Muiswinkel FL, Hack CE, Eikelenboom P. Cytokines associated with amyloid plaques
in Alzheimer's disease brain stimulate human glial and neuronal cell cultures
to secrete early complement proteins, but not C1-inhibitor. Exp Neurol 1999
Nov;160(1):289-99.
(31) Eikelenboom P, van Exel E, Hoozemans JJ,
Veerhuis R, Rozemuller AJ, van Gool WA. Neuroinflammation - an early event
in both the history and pathogenesis of Alzheimer's disease. Neurodegener
Dis 2010;7(1-3):38-41.
(32) Eikelenboom P, Stam FC. Immunoglobulins
and complement factors in senile plaques. An immunoperoxidase study. Acta
Neuropathol 1982;57(2-3):239-42.
(33) D'Andrea MR. Evidence linking neuronal cell
death to autoimmunity in Alzheimer's disease. Brain Res 2003 Aug 22;982(1):19-30.
(34) Veerhuis R, van d, V, Janssen I, Zhan SS,
Van Nostrand WE, Eikelenboom P. Complement activation in amyloid plaques in
Alzheimer's disease brains does not proceed further than C3. Virchows Arch
1995;426(6):603-10.
(35) Veerhuis R, Janssen I, Hack CE, Eikelenboom
P. Early complement components in Alzheimer's disease brains. Acta Neuropathol
1996;91(1):53-60.
(36) Zanjani H, Finch CE, Kemper C, Atkinson
J, McKeel D, Morris JC, et al. Complement activation in very early Alzheimer
disease. Alzheimer Dis Assoc Disord 2005 Apr;19(2):55-66.
(37) Malaguarnera M, Bella R, Alagona G, Ferri
R, Carnemolla A, Pennisi G. Helicobacter pylori and Alzheimer's disease: a
possible link. Eur J Intern Med 2004 Oct;15(6):381-6.
(38) Wozniak MA, Shipley SJ, Combrinck M, Wilcock
GK, Itzhaki RF. Productive herpes simplex virus in brain of elderly normal
subjects and Alzheimer's disease patients. J Med Virol 2005 Feb;75(2):300-6.
(39) Renvoize EB, Awad IO, Hambling MH. A sero-epidemiological
study of conventional infectious agents in Alzheimer's disease. Age Ageing
1987 Sep;16(5):311-4.
(40) Esposito M, Luccarini I, Cicatiello V, De
Falco D, Fiorentini A, Barba P, et al. Immunogenicity and therapeutic efficacy
of phage-displayed beta-amyloid epitopes. Mol Immunol 2008 Feb;45(4):1056-62.
(41) Iacono D, Markesbery WR, Gross M, Pletnikova
O, Rudow G, Zandi P, et al. The Nun study: clinically silent AD, neuronal
hypertrophy, and linguistic skills in early life. Neurology 2009 Sep 1;73(9):665-73.
(42) Scarmeas N, Stern Y, Mayeux R, Luchsinger
JA. Mediterranean diet, Alzheimer disease, and vascular mediation. Arch Neurol
2006 Dec;63(12):1709-17.
(43) van Oijen M, de Jong FJ, Witteman JC, Hofman
A, Koudstaal PJ, Breteler MM. Atherosclerosis and risk for dementia. Ann Neurol
2007 May;61(5):403-10.
(44) de la Torre JC. The vascular hypothesis
of Alzheimer's disease: bench to bedside and beyond. Neurodegener Dis 2010;7(1-3):116-21.
(45) Roe CM, Fitzpatrick AL, Xiong C, Sieh W,
Kuller L, Miller JP, et al. Cancer linked to Alzheimer disease but not vascular
dementia. Neurology 2010 Jan 12;74(2):106-12.
(46) Verreault R, Laurin D, Lindsay J, De Serres
G. Past exposure to vaccines and subsequent risk of Alzheimer's disease. CMAJ
2001 Nov 27;165(11):1495-8.
(47) Garland SM, Smith JS. Human papillomavirus
vaccines: current status and future prospects. Drugs 2010 Jun 18;70(9):1079-98.
(48) Eriksson UK, Gatz M, Dickman PW, Fratiglioni
L, Pedersen NL. Asthma, eczema, rhinitis and the risk for dementia. Dement
Geriatr Cogn Disord 2008;25(2):148-56.
(49) Frecker MF, Pryse-Phillips WE, Strong HR.
Immunological associations in familial and non-familial Alzheimer patients
and their families. Can J Neurol Sci 1994 May;21(2):112-9.
(50) 2010 Alzheimer's disease facts and figures.
Alzheimers Dement 2010 Mar;6(2):158-94.
(51) Seroprevalence of herpes simplex virus type
2 among persons aged 14-49 years--United States, 2005-2008. MMWR Morb Mortal
Wkly Rep 2010 Apr 23;59(15):456-9.
(52) Lin WR, Shang D, Itzhaki RF. Neurotropic
viruses and Alzheimer disease. Interaction of herpes simplex type 1 virus
and apolipoprotein E in the etiology of the disease. Mol Chem Neuropathol
1996 May;28(1-3):135-41.
(53) Esiri MM, Biddolph SC,
Morris CS. Prevalence of Alzheimer plaques in AIDS. J Neurol Neurosurg Psychiatry
1998 Jul;65(1):29-33.
(54) Colucci M, Cammarata S, Assini A, Croce
R, Clerici F, Novello C, et al. The number of pregnancies is a risk factor
for Alzheimer's disease. Eur J Neurol 2006 Dec;13(12):1374-7.
(55) McGeer PL, Rogers
J, McGeer EG. Inflammation, anti-inflammatory agents and Alzheimer disease:
the last 12 years. J Alzheimers Dis 2006;9(3 Suppl):271-6.
(56) Morris MC, Evans DA, Bienias JL, Tangney
CC, Bennett DA, Wilson RS, et al. Consumption of fish and n-3 fatty acids
and risk of incident Alzheimer disease. Arch Neurol 2003 Jul;60(7):940-6.
(57) Laye S. Polyunsaturated fatty acids, neuroinflammation
and well being. Prostaglandins Leukot Essent Fatty Acids 2010 Apr;82(4-6):295-303.
(58) Wolozin B, Kellman W, Ruosseau P, Celesia
GG, Siegel G. Decreased prevalence of Alzheimer disease associated with 3-hydroxy-3-methyglutaryl
coenzyme A reductase inhibitors. Arch Neurol 2000 Oct;57(10):1439-43.
(59) Carter CJ. Convergence of genes implicated
in Alzheimer's disease on the cerebral cholesterol shuttle: APP, cholesterol,
lipoproteins, and atherosclerosis. Neurochem Int 2007 Jan;50(1):12-38.
(60) Bender FC, Whitbeck JC, Ponce dL, Lou H,
Eisenberg RJ, Cohen GH. Specific association of glycoprotein B with lipid
rafts during herpes simplex virus entry. J Virol 2003 Sep;77(17):9542-52.
(61) Carter GC, Bernstone L, Sangani D, Bee JW,
Harder T, James W. HIV entry in macrophages is dependent on intact lipid rafts.
Virology 2009 Mar 30;386(1):192-202.
(62) Weber MS, Stuve O, Neuhaus O, Hartung HP,
Zamvil SS. Spotlight on statins. Int MS J 2007 Sep;14(3):93-7.
(63) Ferreira PC, Piai KA, Takayanagui AM, Segura-Munoz
SI. Aluminum as a risk factor for Alzheimer's disease. Rev Lat Am Enfermagem
2008 Jan;16(1):151-7.
(64) Maton PN, Burton ME.
Antacids revisited: a review of their clinical pharmacology and recommended
therapeutic use. Drugs 1999 Jun;57(6):855-70.
(65) Corder EH, Saunders AM, Strittmatter WJ,
Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type
4 allele and the risk of Alzheimer's disease in late onset families. Science
1993 Aug 13;261(5123):921-3.
(66) Lambert JC, Heath S, Even G, Campion D,
Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants
at CLU and CR1 associated with Alzheimer's disease. Nat Genet 2009 Oct;41(10):1094-9.
(67) Harold D, Abraham R, Hollingworth P, Sims
R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies
variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet
2009 Oct;41(10):1088-93.
(68) Huemer HP, Menzel HJ, Potratz D, Brake B,
Falke D, Utermann G, et al. Herpes simplex virus binds to human serum lipoprotein.
Intervirology 1988;29(2):68-76.
(69) Benga WJ, Krieger SE, Dimitrova M, Zeisel
MB, Parnot M, Lupberger J, et al. Apolipoprotein E interacts with hepatitis
C virus nonstructural protein 5A and determines assembly of infectious particles.
Hepatology 2010 Jan;51(1):43-53.
(70) Kuhlmann I, Minihane AM, Huebbe P, Nebel
A, Rimbach G. Apolipoprotein E genotype and hepatitis C, HIV and herpes simplex
disease risk: a literature review. Lipids Health Dis 2010 Jan 28;9:8.
(71) Male D, Brostoff.J, Roth D, Roitt I. Immunology.
7th ed. Elsevier; 2010.
(72) Powers JH, Buster BL, Reist CJ, Martin E,
Bridges M, Sutherland WM, et al. Complement-independent binding of microorganisms
to primate erythrocytes in vitro by cross-linked monoclonal antibodies via
complement receptor 1. Infect Immun 1995 Apr;63(4):1329-35.
(73) Brunetti CR, Burke RL, Kornfeld S, Gregory
W, Masiarz FR, Dingwell KS, et al. Herpes simplex virus glycoprotein D acquires
mannose 6-phosphate residues and binds to mannose 6-phosphate receptors. J
Biol Chem 1994 Jun 24;269(25):17067-74.
(74) Lemansky P, Brix K, Herzog V. Subcellular
distribution, secretion, and posttranslational modifications of clusterin
in thyrocytes. Exp Cell Res 1999 Aug 25;251(1):147-55.
(75) Tebar F, Bohlander SK, Sorkin A. Clathrin
assembly lymphoid myeloid leukemia (CALM) protein: localization in endocytic-coated
pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated
traffic. Mol Biol Cell 1999 Aug;10(8):2687-702.
(76) Elton D, Simpson-Holley M, Archer K, Medcalf
L, Hallam R, McCauley J, et al. Interaction of the influenza virus nucleoprotein
with the cellular CRM1-mediated nuclear export pathway. J Virol 2001 Jan;75(1):408-19.
(77) Williams P, Verhagen J, Elliott G. Characterization
of a CRM1-dependent nuclear export signal in the C terminus of herpes simplex
virus type 1 tegument protein UL47. J Virol 2008 Nov;82(21):10946-52.
(78) Vecchi M, Polo S, Poupon V, van de Loo JW,
Benmerah A, Di Fiore PP. Nucleocytoplasmic shuttling of endocytic proteins.
J Cell Biol 2001 Jun 25;153(7):1511-7.
(79) Rankl C, Kienberger F, Wildling L, Wruss
J, Gruber HJ, Blaas D, et al. Multiple receptors involved in human rhinovirus
attachment to live cells. Proc Natl Acad Sci U S A 2008 Nov 18;105(46):17778-83.
(80) Martinez WM, Spear PG. Structural features
of nectin-2 (HveB) required for herpes simplex virus entry. J Virol 2001 Nov;75(22):11185-95.
(81) Li Q, Ali MA, Cohen JI. Insulin degrading
enzyme is a cellular receptor mediating varicella-zoster virus infection and
cell-to-cell spread. Cell 2006;127(2):305-16.
(82) Cruts M, van Broeckhoven C. Molecular genetics
of Alzheimer's disease. Ann Med 1998 Dec;30(6):560-5.
(83) Suzuki N, Cheung TT, Cai XD, Odaka A, Otvos
L, Jr., Eckman C, et al. An increased percentage of long amyloid beta protein
secreted by familial amyloid beta protein precursor (beta APP717) mutants.
Science 1994 May 27;264(5163):1336-40.
(84) Citron M, Oltersdorf T, Haass C, McConlogue
L, Hung AY, Seubert P, et al. Mutation of the beta-amyloid precursor protein
in familial Alzheimer's disease increases beta-protein production. Nature
1992 Dec 17;360(6405):672-4.
(85) Chow VW, Mattson MP, Wong PC, Gleichmann
M. An Overview of APP Processing Enzymes and Products. Neuromolecular Med
2009 Nov 24.
(86) Campadelli-Fiume G, Mirandola P, Menotti
L. Human herpesvirus 6: An emerging pathogen. Emerg Infect Dis 1999 May;5(3):353-66.
(87) Kean JM, Rao S, Wang M, Garcea RL. Seroepidemiology
of human polyomaviruses. PLoS Pathog 2009 Mar;5(3):e1000363.
(88) Mast TC, Kierstead L, Gupta SB, Nikas AA,
Kallas EG, Novitsky V, et al. International epidemiology of human pre-existing
adenovirus (Ad) type-5, type-6, type-26 and type-36 neutralizing antibodies:
correlates of high Ad5 titers and implications for potential HIV vaccine trials.
Vaccine 2010 Jan 22;28(4):950-7.
(89) Kokjohn TA, Roher AE. Amyloid precursor
protein transgenic mouse models and Alzheimer's disease: understanding the
paradigms, limitations, and contributions. Alzheimers Dement 2009 Jul;5(4):340-7.
(90) Ebert S, Goos M, Rollwagen L, Baake D, Zech
WD, Esselmann H, et al. Recurrent systemic infections with Streptococcus pneumoniae
do not aggravate the course of experimental neurodegenerative diseases. J
Neurosci Res 2010 Apr;88(5):1124-36.
(91) Stahl T, Reimers C, Johne R, Schliebs R,
Seeger J. Viral-induced inflammation is accompanied by beta-amyloid plaque
reduction in brains of amyloid precursor protein transgenic Tg2576 mice. Eur
J Neurosci 2006 Oct;24(7):1923-34.
(92) Little CS, Hammond CJ, MacIntyre A, Balin
BJ, Appelt DM. Chlamydia pneumoniae induces Alzheimer-like amyloid plaques
in brains of BALB/c mice. Neurobiol Aging 2004 Apr;25(4):419-29.
(93) Slifka MK. The Future of Smallpox Vaccination:
is MVA the key? Med Immunol 2005 Mar 1;4(1):2.
(94) Jenner E. An Inquiry Into the Causes and
Effects of the Variolæ Vaccinæ, Or Cow-Pox. London: 1798.
(95) Schenk D, Barbour R, Dunn W, Gordon G, Grajeda
H, Guido T, et al. Immunization with amyloid-beta attenuates Alzheimer-disease-like
pathology in the PDAPP mouse. Nature 1999 Jul 8;400(6740):173-7.
(96) Roskam S, Neff F, Schwarting R, Bacher M,
Dodel R. APP transgenic mice: the effect of active and passive immunotherapy
in cognitive tasks. Neurosci Biobehav Rev 2010 Mar;34(4):487-99.
(97) Nath A, Hall E, Tuzova M, Dobbs M, Jons
M, Anderson C, et al. Autoantibodies to amyloid beta-peptide (Abeta) are increased
in Alzheimer's disease patients and Abeta antibodies can enhance Abeta neurotoxicity:
implications for disease pathogenesis and vaccine development. Neuromolecular
Med 2003;3(1):29-39.
(98) Das P, Chapoval S, Howard V, David CS, Golde
TE. Immune responses against Abeta1-42 in HLA class II transgenic mice: implications
for Abeta1-42 immune-mediated therapies. Neurobiol Aging 2003 Nov;24(7):969-76.
(99) Ferrer I, Boada RM, Sanchez Guerra ML, Rey
MJ, Costa-Jussa F. Neuropathology and pathogenesis of encephalitis following
amyloid-beta immunization in Alzheimer's disease. Brain Pathol 2004 Jan;14(1):11-20.
(100) Wagner S, Janzen RW, Mohs C, Pohlmann S,
Klingel R, Grutzmacher PW. [Long-term treatment of refractory myasthenia gravis
with immunoadsorption]
Langzeitbehandlung der therapierefraktaren Myasthenia gravis mittels Immunadsorption.
DEP - 20081104. Dtsch Med Wochenschr 2008 Nov;133(46):2377-82.
(101) Belak M, Widder RA, Brunner R, Borberg H,
Haupt WF. Immunoadsorption with protein A sepharose or silica. Lancet 1994
Mar 26;343(8900):792-3.
(102) Yamazaki Z, Idezuki Y, Inoue N, Yoshizawa
H, Yamawaki N, Inagaki K, et al. Extracorporeal immunoadsorption with IM-PH
or IM-TR column. Biomater Artif Cells Artif Organs 1989;17(2):117-24.
(103) Rondeau V, Jacqmin-Gadda H, Commenges D, Helmer C, Dartigues JF. Aluminum
and silica in drinking water and the risk of Alzheimer's disease or cognitive
decline: findings from 15-year follow-up of the PAQUID cohort. Am J Epidemiol
2009 Feb 15;169(4):489-96.
(104) Widner B, Leblhuber F, Walli J, Tilz GP,
Demel U, Fuchs D. Tryptophan degradation and immune activation in Alzheimer's
disease. J Neural Transm 2000;107(3):343-53.
(105) Ravaglia G, Forti P, Maioli F, Bianchi G, Martelli M, Talerico T, et
al. Plasma amino acid concentrations
in patients with amnestic mild cognitive impairment or Alzheimer disease.
Am J Clin Nutr 2004 Aug;80(2):483-8.
(106) Sales C, Oliviero F, Spinella P. [Fish oil
supplementation in rheumatoid arthritis]
La supplementazione dietetica con olio di pesce nell'artrite reumatoide. Reumatismo
2008 Jul;60(3):174-9.
(107) Carter CJ. Interactions between the products
of the Herpes simplex genome and Alzheimer's disease susceptibility genes:
relevance to pathological-signalling cascades. Neurochem Int 2008 May;52(6):920-34.
(108) Carter CJ. Schizophrenia susceptibility genes
directly implicated in the life cycles of pathogens: cytomegalovirus, influenza,
herpes simplex, rubella, and Toxoplasma gondii. Schizophr Bull 2009 Nov;35(6):1163-82.
(109) Bauer PO,
Nukina N. The pathogenic mechanisms of polyglutamine diseases and current
therapeutic strategies. J Neurochem 2009 Sep;110(6):1737-65.
(110) Nukina N. Pathomechanism of polyglutamine
diseases and strategic design for their therapies. Rinsho Shinkeigaku 2008
Nov;48(11):913-4.
(111) Haldane JBS. The origin of life. Rationalist
Annual 148, 3-10. 1928.
Ref Type: Journal (Full)
(112) D'Herelle F. The Bacteriophage; Its Role
in Immunity. Baltimore:
Williams and Wilkins; 1922.
(113) Keller JN. Age-related neuropathology, cognitive
decline, and Alzheimer's disease. Ageing Res Rev 2006 Feb;5(1):1-13.
(114) Brown MG, Vickers IE, Salas RA, Smikle MF.
Seroprevalence of dengue virus antibodies in healthy Jamaicans. Hum Antibodies
2009;18(4):123-6.
(115) Zhang T, Li Y, Ho WZ. Drug abuse, innate
immunity and hepatitis C virus. Rev Med Virol 2006 Sep;16(5):311-27.
(116) Schillinger JA, Xu F, Sternberg MR, Armstrong
GL, Lee FK, Nahmias AJ, et al. National seroprevalence and trends in herpes
simplex virus type 1 in the United States, 1976-1994. Sex Transm Dis 2004
Dec;31(12):753-60.
(117) Hall HI, Song R, Rhodes P, Prejean J, An
Q, Lee LM, et al. Estimation of HIV incidence in the United States. JAMA 2008 Aug 6;300(5):520-9.
(118) Olaleye OD, Omilabu SA, Ilomechina
EN, Fagbami AH. A survey for haemagglutination-inhibiting antibody to
West Nile virus in human and animal sera in Nigeria. Comp Immunol Microbiol Infect
Dis 1990;13(1):35-9.
(119) Schwimmbeck
PL, Dyrberg T, Drachman DB, Oldstone MB. Molecular
mimicry and myasthenia gravis. An autoantigenic site of the acetylcholine
receptor alpha-subunit that has biologic activity and reacts immunochemically
with herpes simplex virus. J Clin Invest 1989 Oct;84(4):1174-80.
(120) Caldarola G, Kneisel A, Hertl M, Feliciani
C. Herpes simplex virus infection in pemphigus vulgaris: clinical and immunological
considerations. Eur J Dermatol 2008 Jul;18(4):440-3.
(121) Mohan A, Chandra S, Agarwal D, Guleria R,
Broor S, Gaur B, et al. Prevalence of viral infection detected by PCR and
RT-PCR in patients with acute exacerbation of COPD: a systematic review. Respirology
2010 Apr;15(3):536-42.
(122) Maghzi AH, Marta M, Bosca I, Etemadifar M,
Dobson R, Maggiore C, et al. Viral pathophysiology of multiple sclerosis:
A role for Epstein-Barr virus infection? Pathophysiology 2010 Jun 8.
(123) Krone B, Pohl D, Rostasy K, Kahler E, Brunner
E, Oeffner F, et al. Common infectious agents in multiple sclerosis: a case-control
study in children. Mult Scler 2008 Jan;14(1):136-9.
(124) Cook
SD, Dowling PC. A possible association
between house pets and multiple sclerosis. Lancet 1977 May 7;1(8019):980-2.
(125) Bansil S, Singhal BS, Ahuja GK, Riise T,
Ladiwala U, Behari M, et al. Multiple sclerosis in India: a case-control study
of environmental exposures. Acta Neurol Scand 1997 Feb;95(2):90-5.
(126) Opsahl ML, Kennedy PG. Investigating the
presence of human herpesvirus 7 and 8 in multiple sclerosis and normal control
brain tissue. J Neurol Sci 2006 Jan 15;240(1-2):37-44.
(127) Jilek S, Jaquiery E, Hirsch HH, Lysandropoulos
A, Canales M, Guignard L, et al. Immune responses to JC virus in patients
with multiple sclerosis treated with natalizumab: a cross-sectional and longitudinal
study. Lancet Neurol 2010 Mar;9(3):264-72.
(128) Andersen O, Lygner PE, Bergstrom T, Andersson
M, Vahlne A. Viral infections trigger multiple sclerosis relapses: a prospective
seroepidemiological study. J Neurol 1993 Jul;240(7):417-22.
(129) Coutinho HD. Burkholderia cepacia complex:
virulence characteristics, importance and relationship with cystic fibrosis.
Indian J Med Sci 2007 Jul;61(7):422-9.


